Chlorine »
PDB 3ilj-3isv »
3iog »
Chlorine in PDB 3iog: Crystal Structure of Cpha N220G Mutant with Inhibitor 18
Enzymatic activity of Crystal Structure of Cpha N220G Mutant with Inhibitor 18
All present enzymatic activity of Crystal Structure of Cpha N220G Mutant with Inhibitor 18:
3.5.2.6;
3.5.2.6;
Protein crystallography data
The structure of Crystal Structure of Cpha N220G Mutant with Inhibitor 18, PDB code: 3iog
was solved by
H.Delbruck,
C.Bebrone,
K.M.V.Hoffmann,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 19.59 / 1.41 |
| Space group | C 2 2 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 42.727, 100.877, 117.559, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 14.3 / 15.5 |
Other elements in 3iog:
The structure of Crystal Structure of Cpha N220G Mutant with Inhibitor 18 also contains other interesting chemical elements:
| Zinc | (Zn) | 1 atom |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Crystal Structure of Cpha N220G Mutant with Inhibitor 18
(pdb code 3iog). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 2 binding sites of Chlorine where determined in the Crystal Structure of Cpha N220G Mutant with Inhibitor 18, PDB code: 3iog:
Jump to Chlorine binding site number: 1; 2;
In total 2 binding sites of Chlorine where determined in the Crystal Structure of Cpha N220G Mutant with Inhibitor 18, PDB code: 3iog:
Jump to Chlorine binding site number: 1; 2;
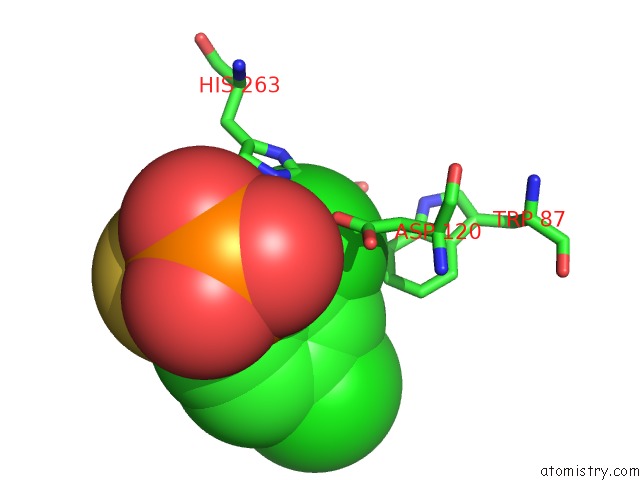
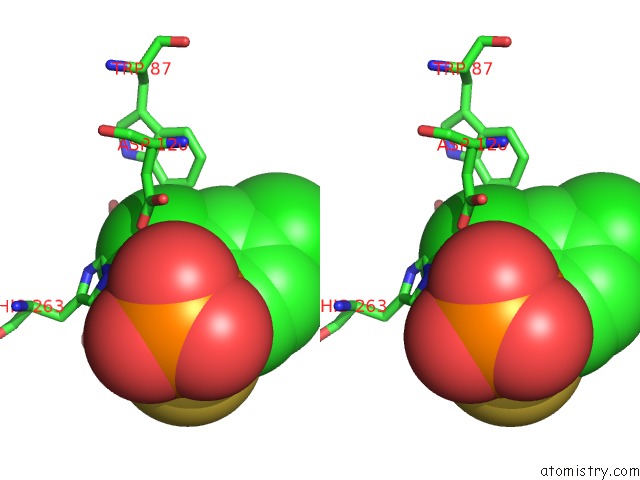
Chlorine binding site 1 out of 2 in 3iog
Go back to
Chlorine binding site 1 out
of 2 in the Crystal Structure of Cpha N220G Mutant with Inhibitor 18
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Crystal Structure of Cpha N220G Mutant with Inhibitor 18 within 5.0Å range:
|
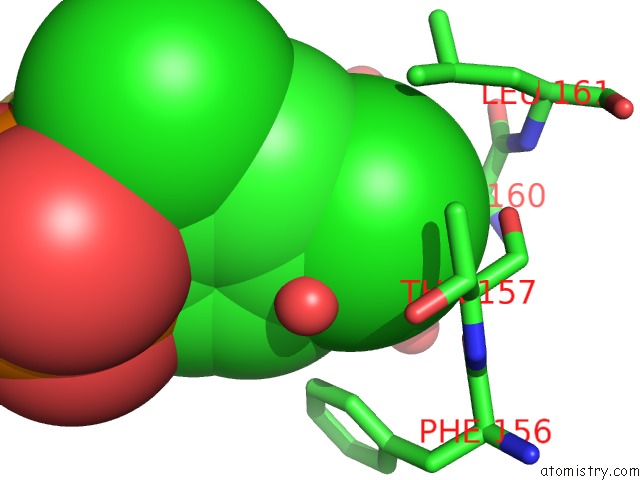
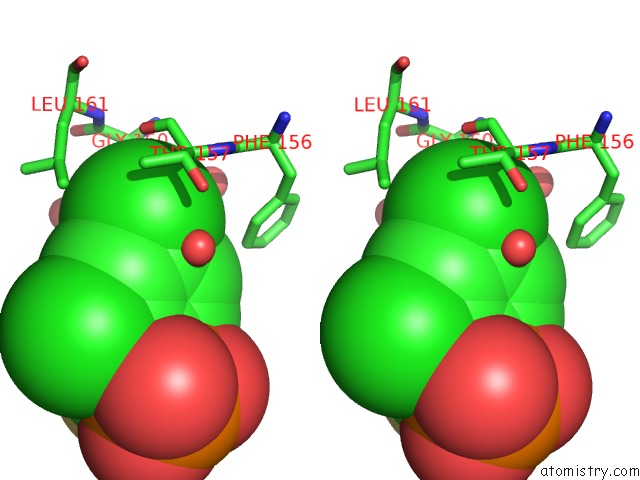
Chlorine binding site 2 out of 2 in 3iog
Go back to
Chlorine binding site 2 out
of 2 in the Crystal Structure of Cpha N220G Mutant with Inhibitor 18
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Crystal Structure of Cpha N220G Mutant with Inhibitor 18 within 5.0Å range:
|
Reference:
P.Lassaux,
M.Hamel,
M.Gulea,
H.Delbruck,
P.S.Mercuri,
L.Horsfall,
D.Dehareng,
M.Kupper,
J.-M.Frere,
K.Hoffmann,
M.Galleni,
C.Bebrone.
Mercaptophosphonate Compounds As Broad-Spectrum Inhibitors of the Metallo-Beta-Lactamases J.Med.Chem. V. 53 4862 2010.
ISSN: ISSN 0022-2623
PubMed: 20527888
DOI: 10.1021/JM100213C
Page generated: Fri Jul 11 06:29:05 2025
ISSN: ISSN 0022-2623
PubMed: 20527888
DOI: 10.1021/JM100213C
Last articles
Mn in 5NRGMn in 5RAA
Mn in 5OMX
Mn in 5OXJ
Mn in 5R7X
Mn in 5OX6
Mn in 5OX5
Mn in 5OR6
Mn in 5OVO
Mn in 5OR2