Chlorine »
PDB 3itd-3jpy »
3ite »
Chlorine in PDB 3ite: The Third Adenylation Domain of the Fungal Sidn Non-Ribosomal Peptide Synthetase
Protein crystallography data
The structure of The Third Adenylation Domain of the Fungal Sidn Non-Ribosomal Peptide Synthetase, PDB code: 3ite
was solved by
T.V.Lee,
J.S.Lott,
R.D.Johnson,
L.J.Johnson,
V.L.Arcus,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 42.62 / 2.00 |
| Space group | P 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 50.598, 75.280, 84.137, 114.85, 94.78, 90.18 |
| R / Rfree (%) | 19.4 / 22.8 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the The Third Adenylation Domain of the Fungal Sidn Non-Ribosomal Peptide Synthetase
(pdb code 3ite). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 2 binding sites of Chlorine where determined in the The Third Adenylation Domain of the Fungal Sidn Non-Ribosomal Peptide Synthetase, PDB code: 3ite:
Jump to Chlorine binding site number: 1; 2;
In total 2 binding sites of Chlorine where determined in the The Third Adenylation Domain of the Fungal Sidn Non-Ribosomal Peptide Synthetase, PDB code: 3ite:
Jump to Chlorine binding site number: 1; 2;
Chlorine binding site 1 out of 2 in 3ite
Go back to
Chlorine binding site 1 out
of 2 in the The Third Adenylation Domain of the Fungal Sidn Non-Ribosomal Peptide Synthetase
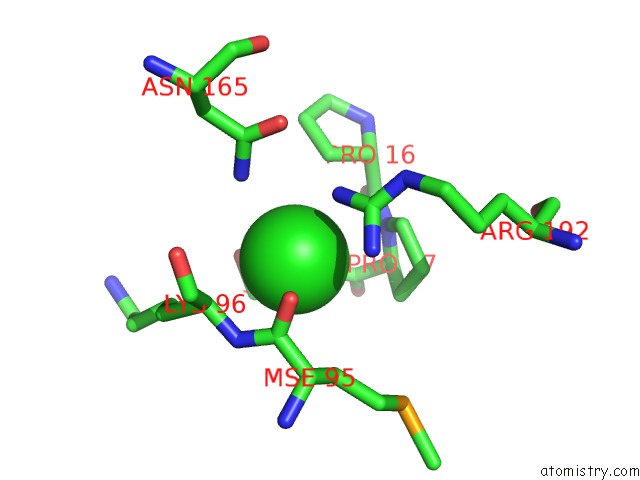
Mono view
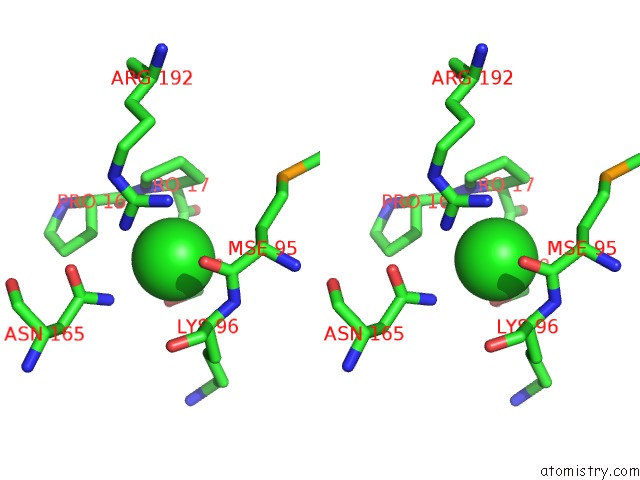
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of The Third Adenylation Domain of the Fungal Sidn Non-Ribosomal Peptide Synthetase within 5.0Å range:
|
Chlorine binding site 2 out of 2 in 3ite
Go back to
Chlorine binding site 2 out
of 2 in the The Third Adenylation Domain of the Fungal Sidn Non-Ribosomal Peptide Synthetase
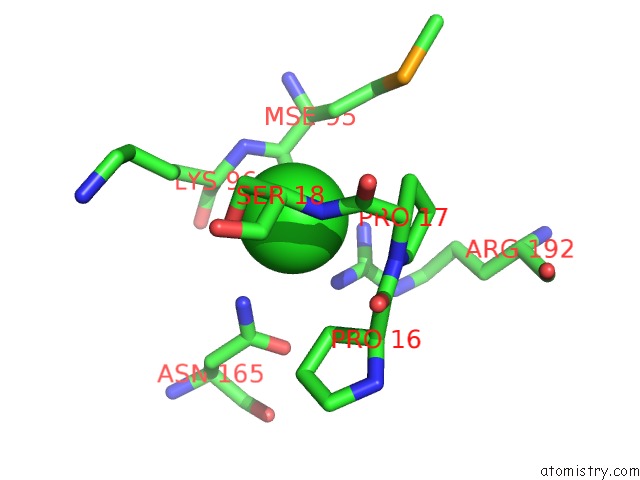
Mono view
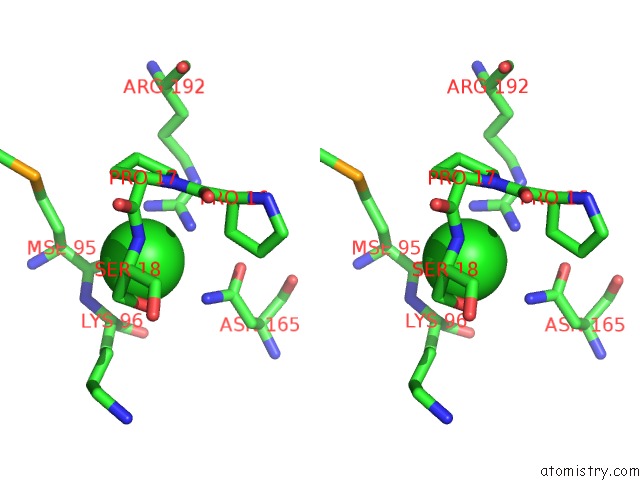
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of The Third Adenylation Domain of the Fungal Sidn Non-Ribosomal Peptide Synthetase within 5.0Å range:
|
Reference:
T.V.Lee,
L.J.Johnson,
R.D.Johnson,
A.Koulman,
G.A.Lane,
J.S.Lott,
V.L.Arcus.
Structure of A Eukaryotic Nonribosomal Peptide Synthetase Adenylation Domain That Activates A Large Hydroxamate Amino Acid in Siderophore Biosynthesis J.Biol.Chem. V. 285 2415 2010.
ISSN: ISSN 0021-9258
PubMed: 19923209
DOI: 10.1074/JBC.M109.071324
Page generated: Fri Jul 11 06:35:43 2025
ISSN: ISSN 0021-9258
PubMed: 19923209
DOI: 10.1074/JBC.M109.071324
Last articles
K in 7QQUK in 7QQV
K in 7QQT
K in 7QQR
K in 7QQS
K in 7QQQ
K in 7QQP
K in 7QQO
K in 7QK5
K in 7QIX