Chlorine »
PDB 3k9u-3khi »
3kdu »
Chlorine in PDB 3kdu: Crystal Structure of Peroxisome Proliferator-Activatedeceptor Alpha (Pparalpha) Complex with N-3-((2-(4-Chlorophenyl)-5-Methyl-1,3- Oxazol-4-Yl)Methoxy)Benzyl)-N-((4-Methylphenoxy)Carbonyl)Glycine
Protein crystallography data
The structure of Crystal Structure of Peroxisome Proliferator-Activatedeceptor Alpha (Pparalpha) Complex with N-3-((2-(4-Chlorophenyl)-5-Methyl-1,3- Oxazol-4-Yl)Methoxy)Benzyl)-N-((4-Methylphenoxy)Carbonyl)Glycine, PDB code: 3kdu
was solved by
J.K.Muckelbauer,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 45.22 / 2.07 |
| Space group | P 41 |
| Cell size a, b, c (Å), α, β, γ (°) | 63.930, 63.930, 126.771, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 17.8 / 23.9 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Crystal Structure of Peroxisome Proliferator-Activatedeceptor Alpha (Pparalpha) Complex with N-3-((2-(4-Chlorophenyl)-5-Methyl-1,3- Oxazol-4-Yl)Methoxy)Benzyl)-N-((4-Methylphenoxy)Carbonyl)Glycine
(pdb code 3kdu). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 2 binding sites of Chlorine where determined in the Crystal Structure of Peroxisome Proliferator-Activatedeceptor Alpha (Pparalpha) Complex with N-3-((2-(4-Chlorophenyl)-5-Methyl-1,3- Oxazol-4-Yl)Methoxy)Benzyl)-N-((4-Methylphenoxy)Carbonyl)Glycine, PDB code: 3kdu:
Jump to Chlorine binding site number: 1; 2;
In total 2 binding sites of Chlorine where determined in the Crystal Structure of Peroxisome Proliferator-Activatedeceptor Alpha (Pparalpha) Complex with N-3-((2-(4-Chlorophenyl)-5-Methyl-1,3- Oxazol-4-Yl)Methoxy)Benzyl)-N-((4-Methylphenoxy)Carbonyl)Glycine, PDB code: 3kdu:
Jump to Chlorine binding site number: 1; 2;
Chlorine binding site 1 out of 2 in 3kdu
Go back to
Chlorine binding site 1 out
of 2 in the Crystal Structure of Peroxisome Proliferator-Activatedeceptor Alpha (Pparalpha) Complex with N-3-((2-(4-Chlorophenyl)-5-Methyl-1,3- Oxazol-4-Yl)Methoxy)Benzyl)-N-((4-Methylphenoxy)Carbonyl)Glycine
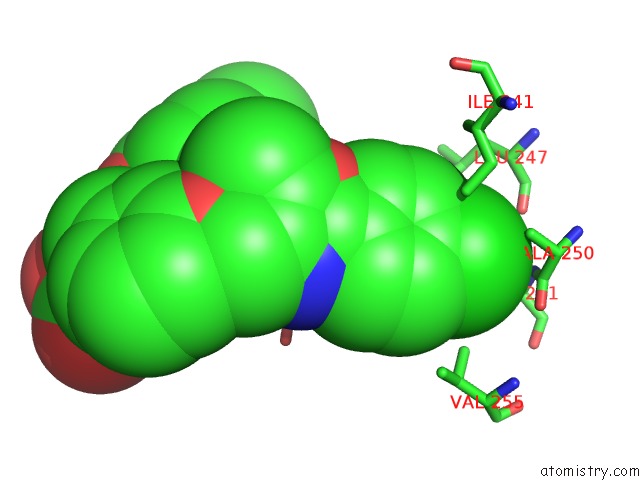
Mono view
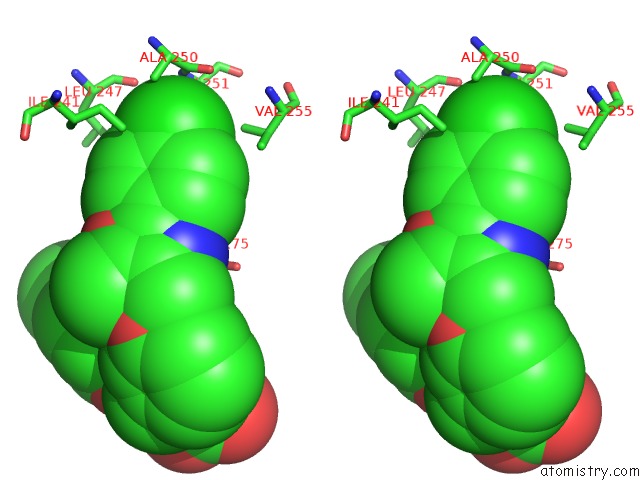
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Crystal Structure of Peroxisome Proliferator-Activatedeceptor Alpha (Pparalpha) Complex with N-3-((2-(4-Chlorophenyl)-5-Methyl-1,3- Oxazol-4-Yl)Methoxy)Benzyl)-N-((4-Methylphenoxy)Carbonyl)Glycine within 5.0Å range:
|
Chlorine binding site 2 out of 2 in 3kdu
Go back to
Chlorine binding site 2 out
of 2 in the Crystal Structure of Peroxisome Proliferator-Activatedeceptor Alpha (Pparalpha) Complex with N-3-((2-(4-Chlorophenyl)-5-Methyl-1,3- Oxazol-4-Yl)Methoxy)Benzyl)-N-((4-Methylphenoxy)Carbonyl)Glycine
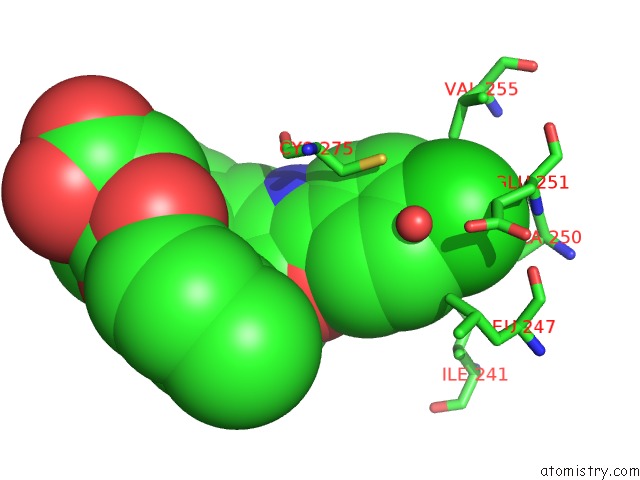
Mono view
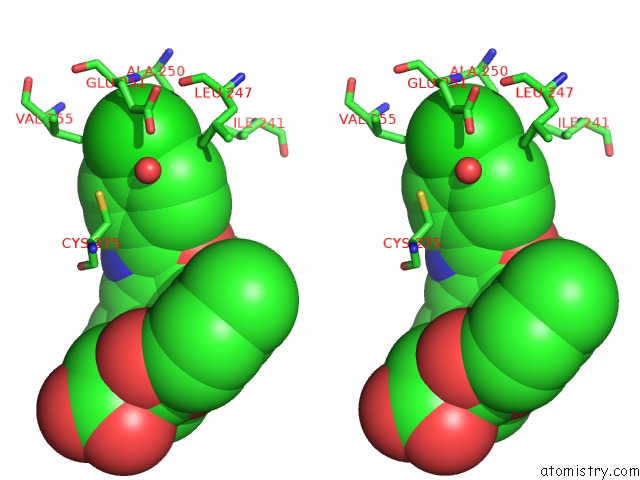
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Crystal Structure of Peroxisome Proliferator-Activatedeceptor Alpha (Pparalpha) Complex with N-3-((2-(4-Chlorophenyl)-5-Methyl-1,3- Oxazol-4-Yl)Methoxy)Benzyl)-N-((4-Methylphenoxy)Carbonyl)Glycine within 5.0Å range:
|
Reference:
J.Li,
L.J.Kennedy,
Y.Shi,
S.Tao,
X.Y.Ye,
S.Y.Chen,
Y.Wang,
A.S.Hernandez,
W.Wang,
P.V.Devasthale,
S.Chen,
Z.Lai,
H.Zhang,
S.Wu,
R.A.Smirk,
S.A.Bolton,
D.E.Ryono,
H.Zhang,
N.K.Lim,
B.C.Chen,
K.T.Locke,
K.M.O'malley,
L.Zhang,
R.A.Srivastava,
B.Miao,
D.S.Meyers,
H.Monshizadegan,
D.Search,
D.Grimm,
R.Zhang,
T.Harrity,
L.K.Kunselman,
M.Cap,
P.Kadiyala,
V.Hosagrahara,
L.Zhang,
C.Xu,
Y.X.Li,
J.K.Muckelbauer,
C.Chang,
Y.An,
S.R.Krystek,
M.A.Blanar,
R.Zahler,
R.Mukherjee,
P.T.Cheng,
J.A.Tino.
Discovery of An Oxybenzylglycine Based Peroxisome Proliferator Activated Receptor Alpha Selective Agonist 2-((3-((2-(4-Chlorophenyl)-5-Methyloxazol-4-Yl)Methoxy) Benzyl)(Methoxycarbonyl)Amino)Acetic Acid (Bms-687453). J.Med.Chem. V. 53 2854 2010.
ISSN: ISSN 0022-2623
PubMed: 20218621
DOI: 10.1021/JM9016812
Page generated: Fri Jul 11 06:57:40 2025
ISSN: ISSN 0022-2623
PubMed: 20218621
DOI: 10.1021/JM9016812
Last articles
Zn in 9QM9Zn in 9S44
Zn in 9OFE
Zn in 9OFC
Zn in 9OFD
Zn in 9OF1
Zn in 9OFB
Zn in 9N0J
Zn in 9M5X
Zn in 9LGI