Chlorine »
PDB 3lnz-3lz0 »
3lpp »
Chlorine in PDB 3lpp: Crystal Complex of N-Terminal Sucrase-Isomaltase with Kotalanol
Enzymatic activity of Crystal Complex of N-Terminal Sucrase-Isomaltase with Kotalanol
All present enzymatic activity of Crystal Complex of N-Terminal Sucrase-Isomaltase with Kotalanol:
3.2.1.10;
3.2.1.10;
Protein crystallography data
The structure of Crystal Complex of N-Terminal Sucrase-Isomaltase with Kotalanol, PDB code: 3lpp
was solved by
L.Sim,
D.R.Rose,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 19.95 / 2.15 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 69.403, 165.763, 341.400, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 17.7 / 22.3 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Crystal Complex of N-Terminal Sucrase-Isomaltase with Kotalanol
(pdb code 3lpp). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total only one binding site of Chlorine was determined in the Crystal Complex of N-Terminal Sucrase-Isomaltase with Kotalanol, PDB code: 3lpp:
In total only one binding site of Chlorine was determined in the Crystal Complex of N-Terminal Sucrase-Isomaltase with Kotalanol, PDB code: 3lpp:
Chlorine binding site 1 out of 1 in 3lpp
Go back to
Chlorine binding site 1 out
of 1 in the Crystal Complex of N-Terminal Sucrase-Isomaltase with Kotalanol
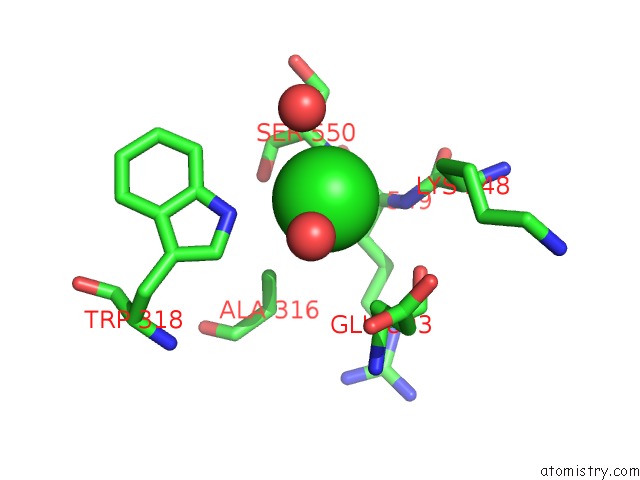
Mono view
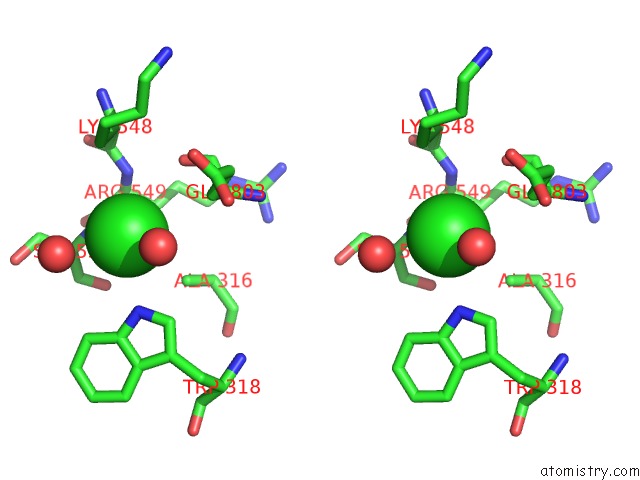
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Crystal Complex of N-Terminal Sucrase-Isomaltase with Kotalanol within 5.0Å range:
|
Reference:
L.Sim,
C.Willemsma,
S.Mohan,
H.Y.Naim,
B.M.Pinto,
D.R.Rose.
Structural Basis For Substrate Selectivity in Human Maltase-Glucoamylase and Sucrase-Isomaltase N-Terminal Domains. J.Biol.Chem. V. 285 17763 2010.
ISSN: ISSN 0021-9258
PubMed: 20356844
DOI: 10.1074/JBC.M109.078980
Page generated: Fri Jul 11 07:33:02 2025
ISSN: ISSN 0021-9258
PubMed: 20356844
DOI: 10.1074/JBC.M109.078980
Last articles
Ir in 6QFUIr in 6DLR
Ir in 6PLB
Ir in 6LXX
Ir in 6GMI
Ir in 6JQ6
Ir in 6ESU
Ir in 6ESS
Ir in 6E80
Ir in 6D3P