Chlorine »
PDB 3qvp-3r40 »
3qzu »
Chlorine in PDB 3qzu: Crystal Structure of Bacillus Subtilis Lipase A 7-Fold Mutant; the Outcome of Directed Evolution Towards Thermostability
Enzymatic activity of Crystal Structure of Bacillus Subtilis Lipase A 7-Fold Mutant; the Outcome of Directed Evolution Towards Thermostability
All present enzymatic activity of Crystal Structure of Bacillus Subtilis Lipase A 7-Fold Mutant; the Outcome of Directed Evolution Towards Thermostability:
3.1.1.3;
3.1.1.3;
Protein crystallography data
The structure of Crystal Structure of Bacillus Subtilis Lipase A 7-Fold Mutant; the Outcome of Directed Evolution Towards Thermostability, PDB code: 3qzu
was solved by
T.Pijning,
W.Augustyniak,
M.T.Reetz,
B.W.Dijkstra,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 76.16 / 1.85 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 59.163, 45.511, 77.613, 90.00, 101.10, 90.00 |
| R / Rfree (%) | 17.3 / 22.3 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Crystal Structure of Bacillus Subtilis Lipase A 7-Fold Mutant; the Outcome of Directed Evolution Towards Thermostability
(pdb code 3qzu). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 2 binding sites of Chlorine where determined in the Crystal Structure of Bacillus Subtilis Lipase A 7-Fold Mutant; the Outcome of Directed Evolution Towards Thermostability, PDB code: 3qzu:
Jump to Chlorine binding site number: 1; 2;
In total 2 binding sites of Chlorine where determined in the Crystal Structure of Bacillus Subtilis Lipase A 7-Fold Mutant; the Outcome of Directed Evolution Towards Thermostability, PDB code: 3qzu:
Jump to Chlorine binding site number: 1; 2;
Chlorine binding site 1 out of 2 in 3qzu
Go back to
Chlorine binding site 1 out
of 2 in the Crystal Structure of Bacillus Subtilis Lipase A 7-Fold Mutant; the Outcome of Directed Evolution Towards Thermostability
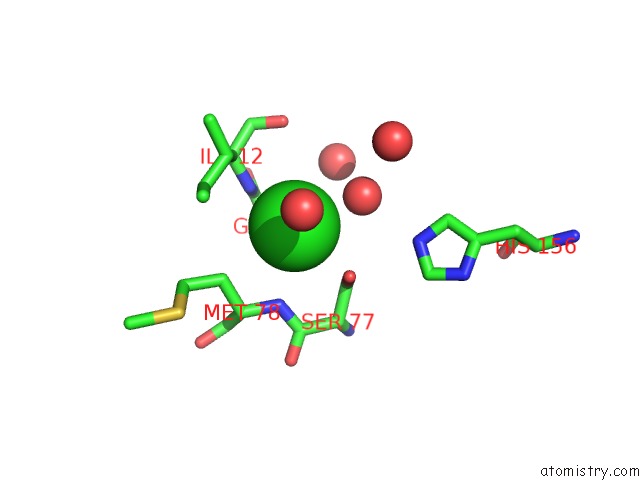
Mono view
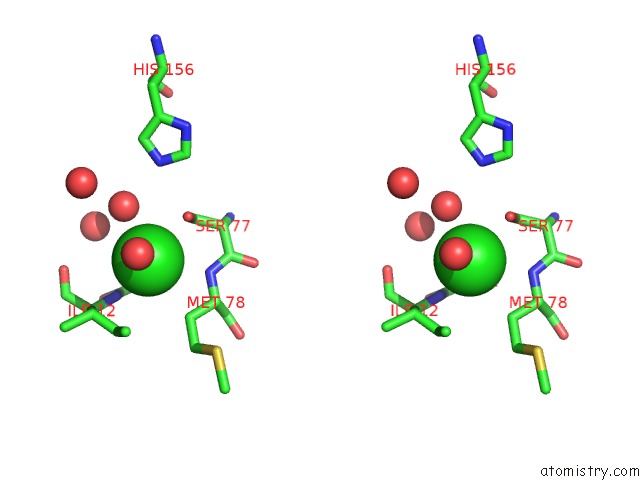
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Crystal Structure of Bacillus Subtilis Lipase A 7-Fold Mutant; the Outcome of Directed Evolution Towards Thermostability within 5.0Å range:
|
Chlorine binding site 2 out of 2 in 3qzu
Go back to
Chlorine binding site 2 out
of 2 in the Crystal Structure of Bacillus Subtilis Lipase A 7-Fold Mutant; the Outcome of Directed Evolution Towards Thermostability
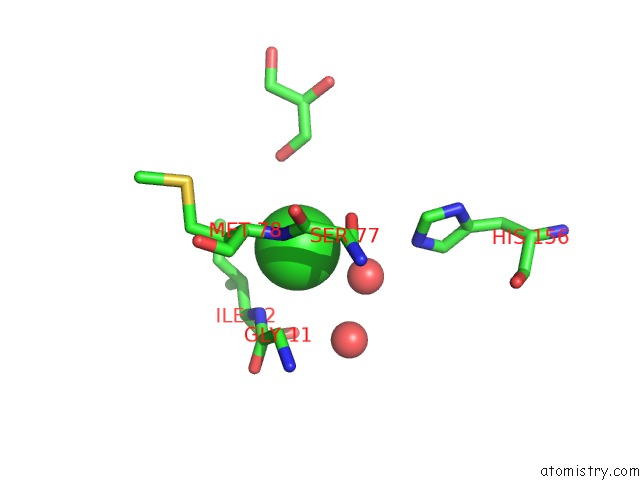
Mono view
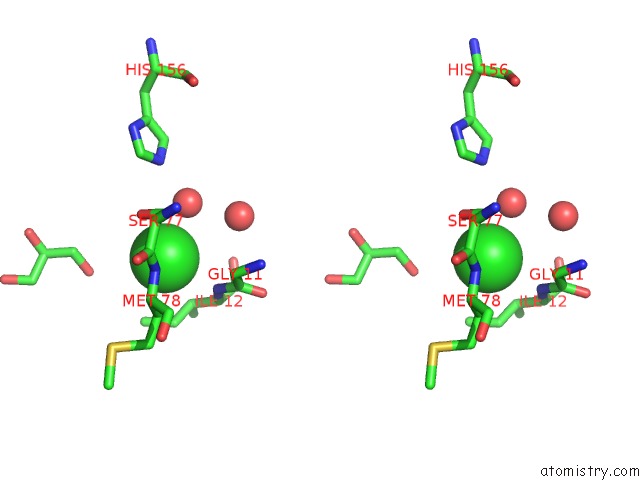
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Crystal Structure of Bacillus Subtilis Lipase A 7-Fold Mutant; the Outcome of Directed Evolution Towards Thermostability within 5.0Å range:
|
Reference:
W.Augustyniak,
A.A.Brzezinska,
T.Pijning,
H.Wienk,
R.Boelens,
B.W.Dijkstra,
M.T.Reetz.
Biophysical Characterization of Mutants of Bacillus Subtilis Lipase Evolved For Thermostability: Factors Contributing to Increased Activity Retention. Protein Sci. V. 21 487 2012.
ISSN: ISSN 0961-8368
PubMed: 22267088
DOI: 10.1002/PRO.2031
Page generated: Fri Jul 11 09:38:05 2025
ISSN: ISSN 0961-8368
PubMed: 22267088
DOI: 10.1002/PRO.2031
Last articles
K in 8R6KK in 8R56
K in 8QZ3
K in 8R37
K in 8QXV
K in 8QXU
K in 8QZ2
K in 8QZ1
K in 8QXT
K in 8QXS