Chlorine »
PDB 3rmn-3ruu »
3roa »
Chlorine in PDB 3roa: Candida Glabrata Dihydrofolate Reductase Complexed with Nadph and 6- Ethyl-5-[(3R)-3-[3-Methoxy-5-(Morpholin-4-Yl)Phenyl]But-1-Yn-1- Yl]Pyrimidine-2,4-Diamine (UCP1004)
Protein crystallography data
The structure of Candida Glabrata Dihydrofolate Reductase Complexed with Nadph and 6- Ethyl-5-[(3R)-3-[3-Methoxy-5-(Morpholin-4-Yl)Phenyl]But-1-Yn-1- Yl]Pyrimidine-2,4-Diamine (UCP1004), PDB code: 3roa
was solved by
J.L.Paulsen,
A.C.Anderson,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 42.72 / 2.30 |
| Space group | P 41 |
| Cell size a, b, c (Å), α, β, γ (°) | 42.710, 42.710, 229.470, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 20.4 / 25.8 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Candida Glabrata Dihydrofolate Reductase Complexed with Nadph and 6- Ethyl-5-[(3R)-3-[3-Methoxy-5-(Morpholin-4-Yl)Phenyl]But-1-Yn-1- Yl]Pyrimidine-2,4-Diamine (UCP1004)
(pdb code 3roa). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 2 binding sites of Chlorine where determined in the Candida Glabrata Dihydrofolate Reductase Complexed with Nadph and 6- Ethyl-5-[(3R)-3-[3-Methoxy-5-(Morpholin-4-Yl)Phenyl]But-1-Yn-1- Yl]Pyrimidine-2,4-Diamine (UCP1004), PDB code: 3roa:
Jump to Chlorine binding site number: 1; 2;
In total 2 binding sites of Chlorine where determined in the Candida Glabrata Dihydrofolate Reductase Complexed with Nadph and 6- Ethyl-5-[(3R)-3-[3-Methoxy-5-(Morpholin-4-Yl)Phenyl]But-1-Yn-1- Yl]Pyrimidine-2,4-Diamine (UCP1004), PDB code: 3roa:
Jump to Chlorine binding site number: 1; 2;
Chlorine binding site 1 out of 2 in 3roa
Go back to
Chlorine binding site 1 out
of 2 in the Candida Glabrata Dihydrofolate Reductase Complexed with Nadph and 6- Ethyl-5-[(3R)-3-[3-Methoxy-5-(Morpholin-4-Yl)Phenyl]But-1-Yn-1- Yl]Pyrimidine-2,4-Diamine (UCP1004)
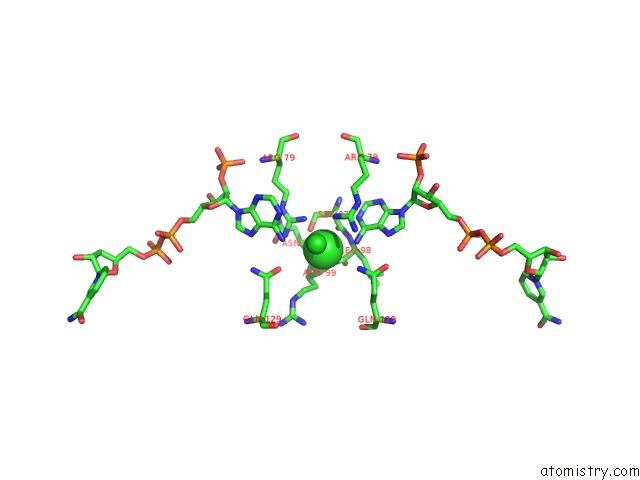
Mono view
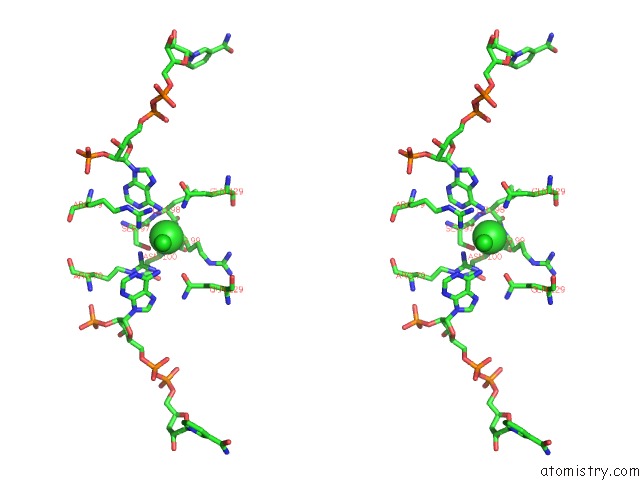
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Candida Glabrata Dihydrofolate Reductase Complexed with Nadph and 6- Ethyl-5-[(3R)-3-[3-Methoxy-5-(Morpholin-4-Yl)Phenyl]But-1-Yn-1- Yl]Pyrimidine-2,4-Diamine (UCP1004) within 5.0Å range:
|
Chlorine binding site 2 out of 2 in 3roa
Go back to
Chlorine binding site 2 out
of 2 in the Candida Glabrata Dihydrofolate Reductase Complexed with Nadph and 6- Ethyl-5-[(3R)-3-[3-Methoxy-5-(Morpholin-4-Yl)Phenyl]But-1-Yn-1- Yl]Pyrimidine-2,4-Diamine (UCP1004)
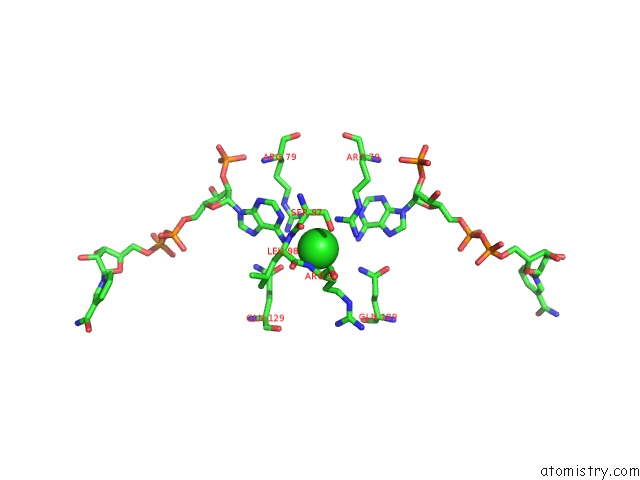
Mono view
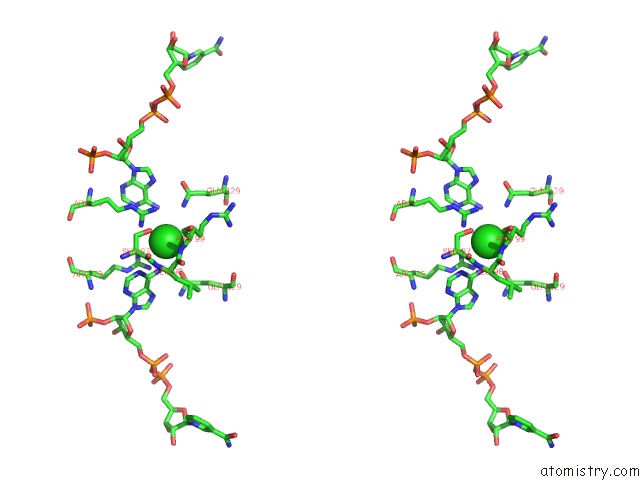
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Candida Glabrata Dihydrofolate Reductase Complexed with Nadph and 6- Ethyl-5-[(3R)-3-[3-Methoxy-5-(Morpholin-4-Yl)Phenyl]But-1-Yn-1- Yl]Pyrimidine-2,4-Diamine (UCP1004) within 5.0Å range:
|
Reference:
J.L.Paulsen,
K.Viswanathan,
D.L.Wright,
A.C.Anderson.
Structural Analysis of the Active Sites of Dihydrofolate Reductase From Two Species of Candida Uncovers Ligand-Induced Conformational Changes Shared Among Species. Bioorg.Med.Chem.Lett. V. 23 1279 2013.
ISSN: ISSN 0960-894X
PubMed: 23375226
DOI: 10.1016/J.BMCL.2013.01.008
Page generated: Fri Jul 11 09:55:31 2025
ISSN: ISSN 0960-894X
PubMed: 23375226
DOI: 10.1016/J.BMCL.2013.01.008
Last articles
Na in 2BY4Na in 2BWU
Na in 2BL2
Na in 2BS4
Na in 2BS3
Na in 2BS2
Na in 2BQO
Na in 2BQN
Na in 2BQL
Na in 2BQJ