Chlorine »
PDB 3u6a-3ue5 »
3u78 »
Chlorine in PDB 3u78: E67-2 Selectively Inhibits KIAA1718, A Human Histone H3 Lysine 9 Jumonji Demethylase
Protein crystallography data
The structure of E67-2 Selectively Inhibits KIAA1718, A Human Histone H3 Lysine 9 Jumonji Demethylase, PDB code: 3u78
was solved by
A.K.Upadhyay,
X.Cheng,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 26.59 / 2.69 |
| Space group | P 61 2 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 77.429, 77.429, 289.685, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 19.8 / 26.5 |
Other elements in 3u78:
The structure of E67-2 Selectively Inhibits KIAA1718, A Human Histone H3 Lysine 9 Jumonji Demethylase also contains other interesting chemical elements:
| Nickel | (Ni) | 1 atom |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the E67-2 Selectively Inhibits KIAA1718, A Human Histone H3 Lysine 9 Jumonji Demethylase
(pdb code 3u78). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total only one binding site of Chlorine was determined in the E67-2 Selectively Inhibits KIAA1718, A Human Histone H3 Lysine 9 Jumonji Demethylase, PDB code: 3u78:
In total only one binding site of Chlorine was determined in the E67-2 Selectively Inhibits KIAA1718, A Human Histone H3 Lysine 9 Jumonji Demethylase, PDB code: 3u78:
Chlorine binding site 1 out of 1 in 3u78
Go back to
Chlorine binding site 1 out
of 1 in the E67-2 Selectively Inhibits KIAA1718, A Human Histone H3 Lysine 9 Jumonji Demethylase
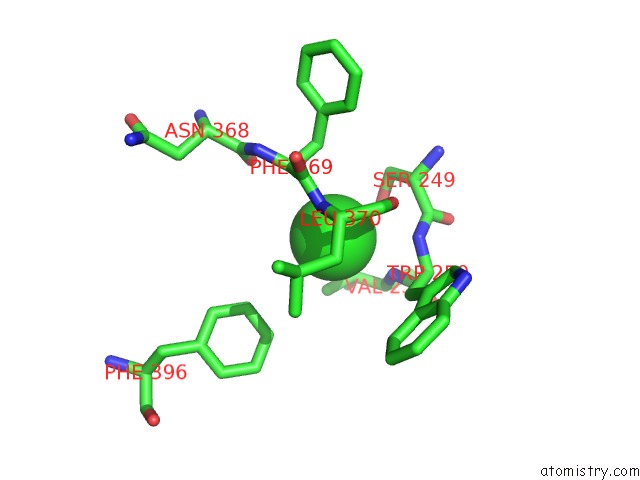
Mono view
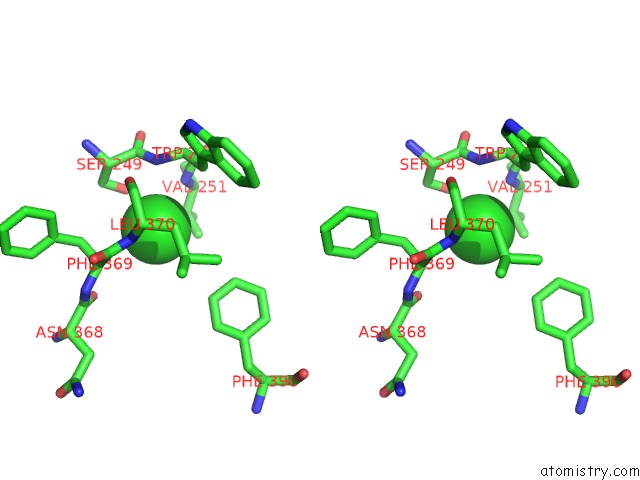
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of E67-2 Selectively Inhibits KIAA1718, A Human Histone H3 Lysine 9 Jumonji Demethylase within 5.0Å range:
|
Reference:
A.K.Upadhyay,
D.Rotili,
J.W.Han,
R.Hu,
Y.Chang,
D.Labella,
X.Zhang,
Y.S.Yoon,
A.Mai,
X.Cheng.
An Analog of Bix-01294 Selectively Inhibits A Family of Histone H3 Lysine 9 Jumonji Demethylases. J.Mol.Biol. V. 416 319 2012.
ISSN: ISSN 0022-2836
PubMed: 22227394
DOI: 10.1016/J.JMB.2011.12.036
Page generated: Fri Jul 11 11:07:41 2025
ISSN: ISSN 0022-2836
PubMed: 22227394
DOI: 10.1016/J.JMB.2011.12.036
Last articles
Fe in 7DIOFe in 7DIH
Fe in 7DI8
Fe in 7DI3
Fe in 7DGM
Fe in 7DGN
Fe in 7DGO
Fe in 7DGL
Fe in 7DE4
Fe in 7DEG