Chlorine »
PDB 3ue8-3unh »
3ugv »
Chlorine in PDB 3ugv: Crystal Structure of An Enolase From Alpha Pretobacterium BAL199 (Efi Target Efi-501650) with Bound Mg
Protein crystallography data
The structure of Crystal Structure of An Enolase From Alpha Pretobacterium BAL199 (Efi Target Efi-501650) with Bound Mg, PDB code: 3ugv
was solved by
M.W.Vetting,
R.Toro,
R.Bhosle,
S.R.Wasserman,
L.L.Morisco,
B.Hillerich,
E.Washington,
A.Scott Glenn,
S.Chowdhury,
B.Evans,
J.Hammonds,
W.D.Zencheck,
H.J.Imker,
J.A.Gerlt,
S.C.Almo,
Enzyme Function Initiative(Efi),
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 114.64 / 2.30 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 152.711, 152.984, 173.535, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 16.1 / 20.8 |
Other elements in 3ugv:
The structure of Crystal Structure of An Enolase From Alpha Pretobacterium BAL199 (Efi Target Efi-501650) with Bound Mg also contains other interesting chemical elements:
| Nickel | (Ni) | 2 atoms |
| Magnesium | (Mg) | 16 atoms |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Crystal Structure of An Enolase From Alpha Pretobacterium BAL199 (Efi Target Efi-501650) with Bound Mg
(pdb code 3ugv). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 3 binding sites of Chlorine where determined in the Crystal Structure of An Enolase From Alpha Pretobacterium BAL199 (Efi Target Efi-501650) with Bound Mg, PDB code: 3ugv:
Jump to Chlorine binding site number: 1; 2; 3;
In total 3 binding sites of Chlorine where determined in the Crystal Structure of An Enolase From Alpha Pretobacterium BAL199 (Efi Target Efi-501650) with Bound Mg, PDB code: 3ugv:
Jump to Chlorine binding site number: 1; 2; 3;
Chlorine binding site 1 out of 3 in 3ugv
Go back to
Chlorine binding site 1 out
of 3 in the Crystal Structure of An Enolase From Alpha Pretobacterium BAL199 (Efi Target Efi-501650) with Bound Mg
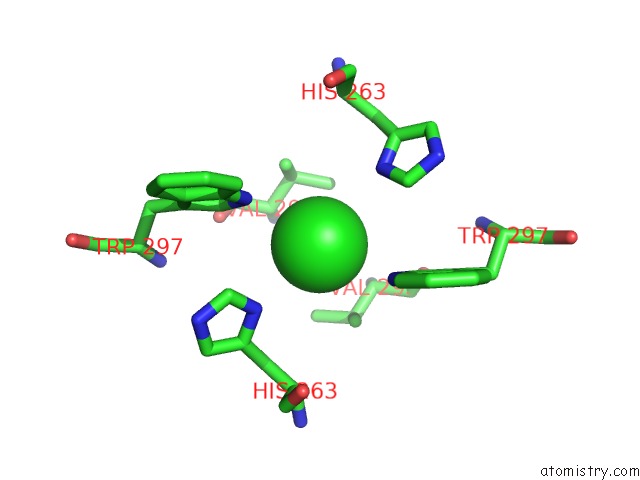
Mono view
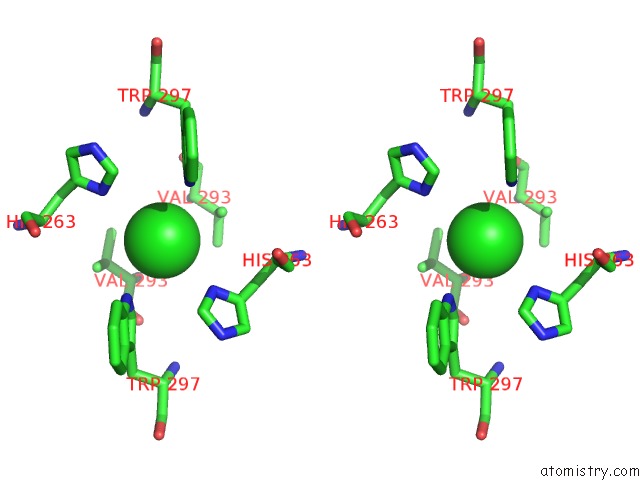
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Crystal Structure of An Enolase From Alpha Pretobacterium BAL199 (Efi Target Efi-501650) with Bound Mg within 5.0Å range:
|
Chlorine binding site 2 out of 3 in 3ugv
Go back to
Chlorine binding site 2 out
of 3 in the Crystal Structure of An Enolase From Alpha Pretobacterium BAL199 (Efi Target Efi-501650) with Bound Mg
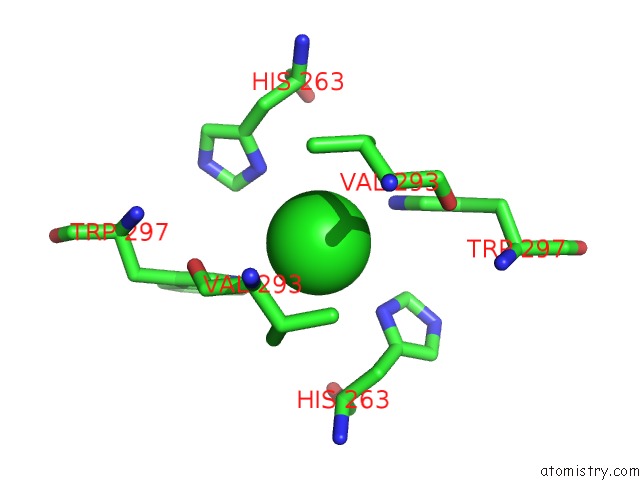
Mono view
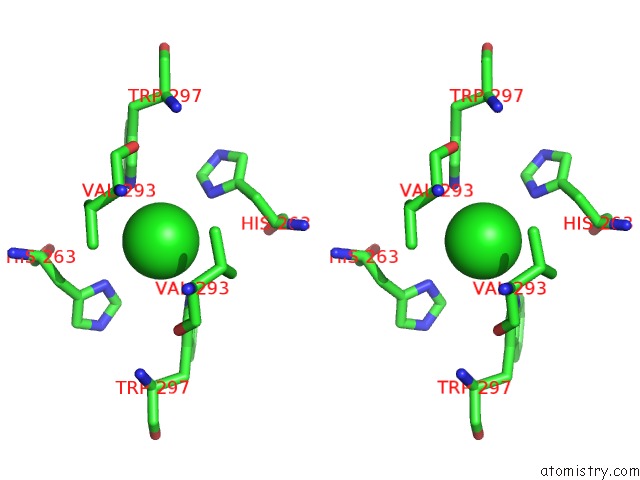
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Crystal Structure of An Enolase From Alpha Pretobacterium BAL199 (Efi Target Efi-501650) with Bound Mg within 5.0Å range:
|
Chlorine binding site 3 out of 3 in 3ugv
Go back to
Chlorine binding site 3 out
of 3 in the Crystal Structure of An Enolase From Alpha Pretobacterium BAL199 (Efi Target Efi-501650) with Bound Mg
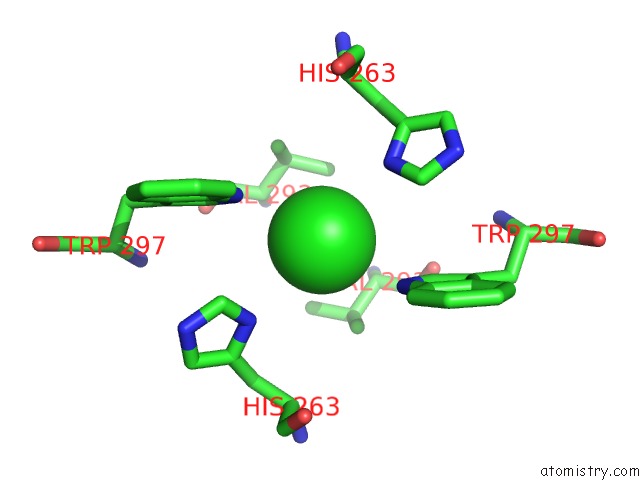
Mono view
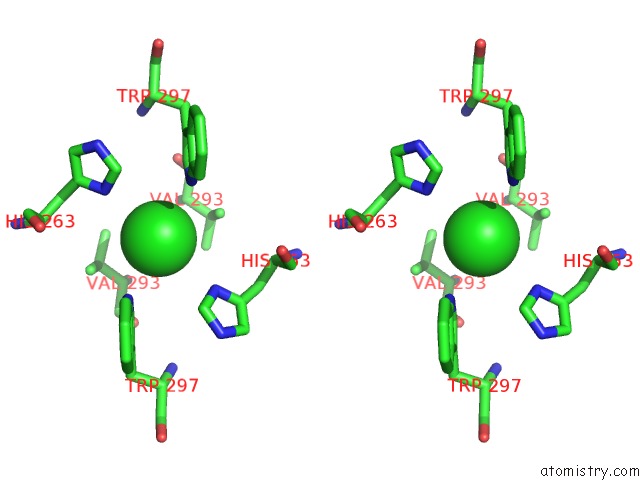
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Crystal Structure of An Enolase From Alpha Pretobacterium BAL199 (Efi Target Efi-501650) with Bound Mg within 5.0Å range:
|
Reference:
M.W.Vetting,
R.Toro,
R.Bhosle,
S.R.Wasserman,
L.L.Morisco,
B.Hillerich,
E.Washington,
A.Scott Glenn,
S.Chowdhury,
B.Evans,
J.Hammonds,
W.D.Zencheck,
H.J.Imker,
J.A.Gerlt,
S.C.Almo,
Enzyme Function Initiative (Efi).
Crystal Structure of An Enolase From Alpha Pretobacterium BAL199 (Efi Target Efi-501650) with Bound Mg To Be Published.
Page generated: Fri Jul 11 11:15:41 2025
Last articles
Mg in 6UBZMg in 6UBY
Mg in 6U9H
Mg in 6UBR
Mg in 6UBN
Mg in 6UBQ
Mg in 6U9Q
Mg in 6U91
Mg in 6U8Q
Mg in 6U8W