Chlorine »
PDB 3zot-3zx2 »
3zs0 »
Chlorine in PDB 3zs0: Human Myeloperoxidase Inactivated By TX2
Enzymatic activity of Human Myeloperoxidase Inactivated By TX2
All present enzymatic activity of Human Myeloperoxidase Inactivated By TX2:
1.11.2.2;
1.11.2.2;
Protein crystallography data
The structure of Human Myeloperoxidase Inactivated By TX2, PDB code: 3zs0
was solved by
A.K.Tiden,
T.Sjogren,
M.Svensson,
A.Bernlind,
R.Senthilmohan,
F.Auchere,
H.Norman,
P.O.Markgren,
S.Gustavsson,
S.Schmidt,
S.Lundquist,
L.V.Forbes,
N.J.Magon,
G.N.Jameson,
H.Eriksson,
A.J.Kettle,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 30.00 / 2.30 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 93.905, 64.111, 111.452, 90.00, 97.12, 90.00 |
| R / Rfree (%) | 17 / 20.7 |
Other elements in 3zs0:
The structure of Human Myeloperoxidase Inactivated By TX2 also contains other interesting chemical elements:
| Fluorine | (F) | 2 atoms |
| Iron | (Fe) | 2 atoms |
| Calcium | (Ca) | 2 atoms |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Human Myeloperoxidase Inactivated By TX2
(pdb code 3zs0). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 2 binding sites of Chlorine where determined in the Human Myeloperoxidase Inactivated By TX2, PDB code: 3zs0:
Jump to Chlorine binding site number: 1; 2;
In total 2 binding sites of Chlorine where determined in the Human Myeloperoxidase Inactivated By TX2, PDB code: 3zs0:
Jump to Chlorine binding site number: 1; 2;
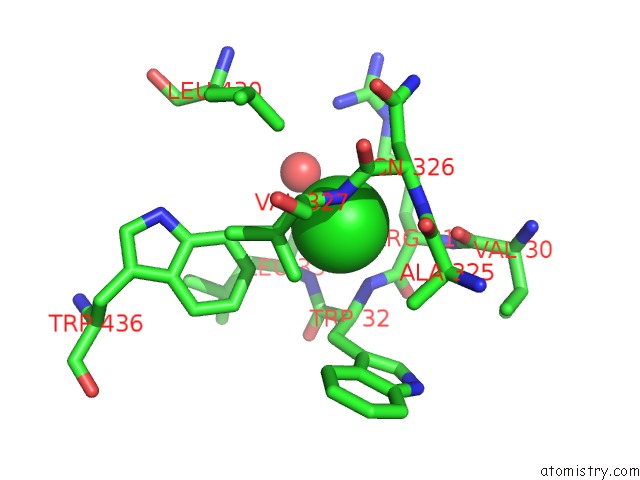
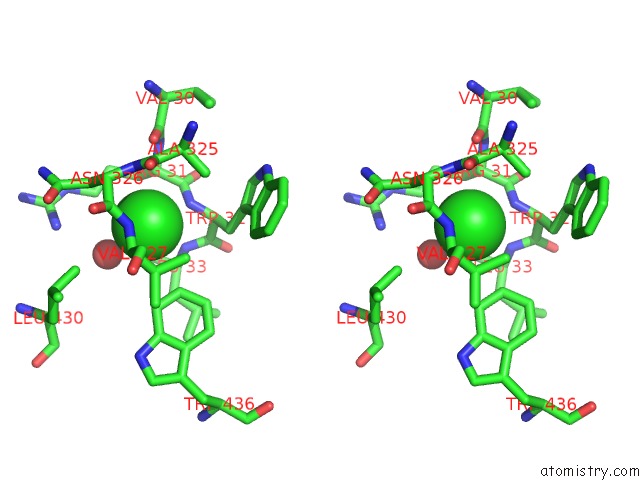
Chlorine binding site 1 out of 2 in 3zs0
Go back to
Chlorine binding site 1 out
of 2 in the Human Myeloperoxidase Inactivated By TX2
Mono view
Stereo pair view

Mono view

Stereo pair view
|
|
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Human Myeloperoxidase Inactivated By TX2 within 5.0Å range:
|
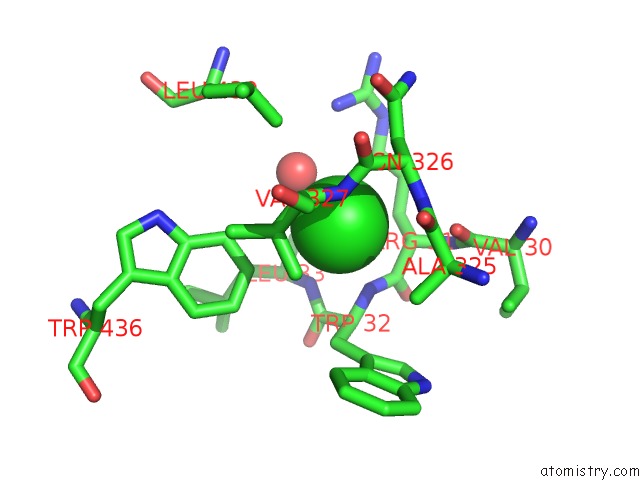
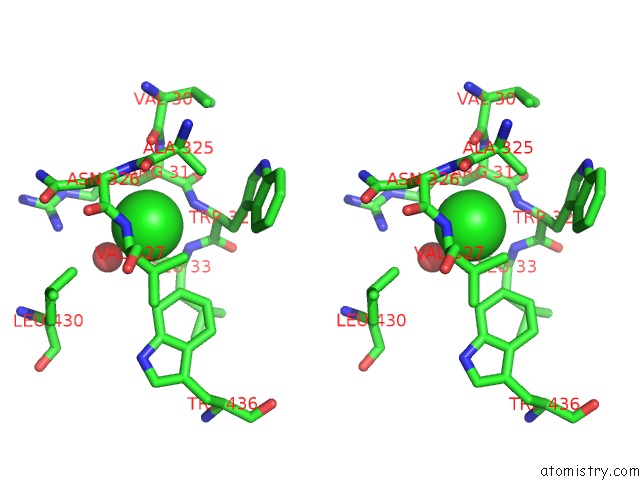
Chlorine binding site 2 out of 2 in 3zs0
Go back to
Chlorine binding site 2 out
of 2 in the Human Myeloperoxidase Inactivated By TX2
Mono view
Stereo pair view

Mono view

Stereo pair view
|
|
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Human Myeloperoxidase Inactivated By TX2 within 5.0Å range:
|
Reference:
A.K.Tiden,
T.Sjogren,
M.Svensson,
A.Bernlind,
R.Senthilmohan,
F.Auchere,
H.Norman,
P.O.Markgren,
S.Gustavsson,
S.Schmidt,
S.Lundquist,
L.V.Forbes,
N.J.Magon,
L.N.Paton,
G.N.Jameson,
H.Eriksson,
A.J.Kettle.
2-Thioxanthines Are Mechanism-Based Inactivators of Myeloperoxidase That Block Oxidative Stress During Inflammation. J.Biol.Chem. V. 286 37578 2011.
ISSN: ISSN 0021-9258
PubMed: 21880720
DOI: 10.1074/JBC.M111.266981
Page generated: Fri Jul 11 12:29:01 2025
ISSN: ISSN 0021-9258
PubMed: 21880720
DOI: 10.1074/JBC.M111.266981
Last articles
W in 2E7ZW in 2HCM
W in 1I95
W in 2AKC
W in 2E6B
W in 1Z26
W in 1Z7M
W in 1N7D
W in 1YTW
W in 1XDY