Chlorine »
PDB 3zx3-4a7i »
4a25 »
Chlorine in PDB 4a25: X-Ray Structure Dps From Kineococcus Radiotolerans in Complex with Mn (II) Ions.
Protein crystallography data
The structure of X-Ray Structure Dps From Kineococcus Radiotolerans in Complex with Mn (II) Ions., PDB code: 4a25
was solved by
A.Ilari,
A.Fiorillo,
M.Ardini,
S.Stefanini,
E.Chiancone,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 75.24 / 2.00 |
| Space group | H 3 |
| Cell size a, b, c (Å), α, β, γ (°) | 150.474, 150.474, 86.618, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 19.42 / 23.226 |
Other elements in 4a25:
The structure of X-Ray Structure Dps From Kineococcus Radiotolerans in Complex with Mn (II) Ions. also contains other interesting chemical elements:
| Manganese | (Mn) | 10 atoms |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the X-Ray Structure Dps From Kineococcus Radiotolerans in Complex with Mn (II) Ions.
(pdb code 4a25). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 6 binding sites of Chlorine where determined in the X-Ray Structure Dps From Kineococcus Radiotolerans in Complex with Mn (II) Ions., PDB code: 4a25:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5; 6;
In total 6 binding sites of Chlorine where determined in the X-Ray Structure Dps From Kineococcus Radiotolerans in Complex with Mn (II) Ions., PDB code: 4a25:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5; 6;
Chlorine binding site 1 out of 6 in 4a25
Go back to
Chlorine binding site 1 out
of 6 in the X-Ray Structure Dps From Kineococcus Radiotolerans in Complex with Mn (II) Ions.
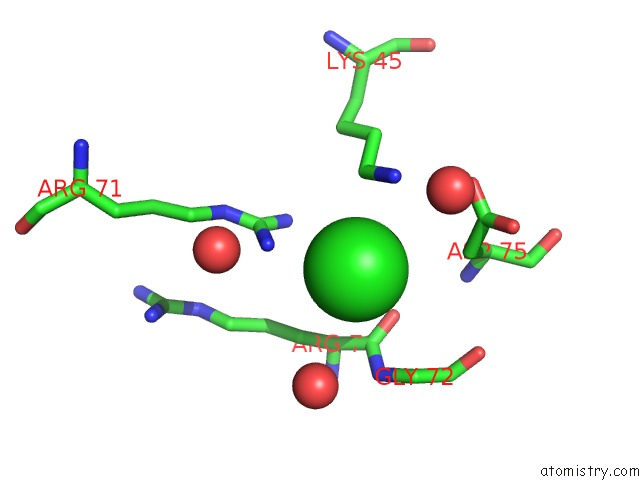
Mono view
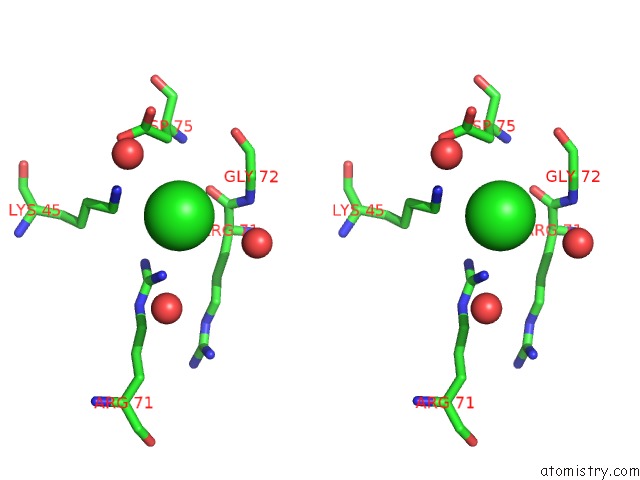
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of X-Ray Structure Dps From Kineococcus Radiotolerans in Complex with Mn (II) Ions. within 5.0Å range:
|
Chlorine binding site 2 out of 6 in 4a25
Go back to
Chlorine binding site 2 out
of 6 in the X-Ray Structure Dps From Kineococcus Radiotolerans in Complex with Mn (II) Ions.
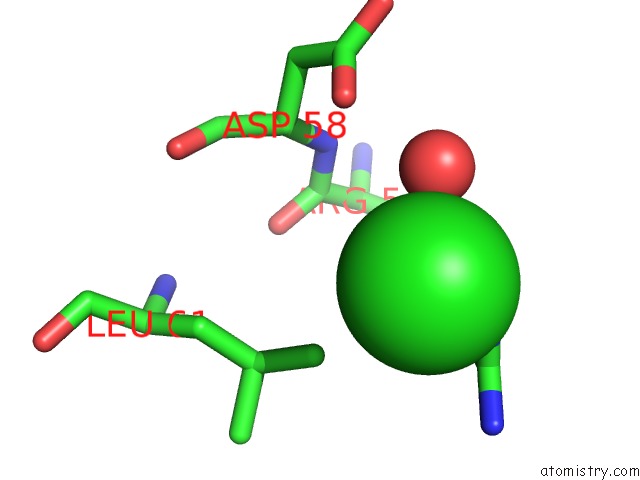
Mono view
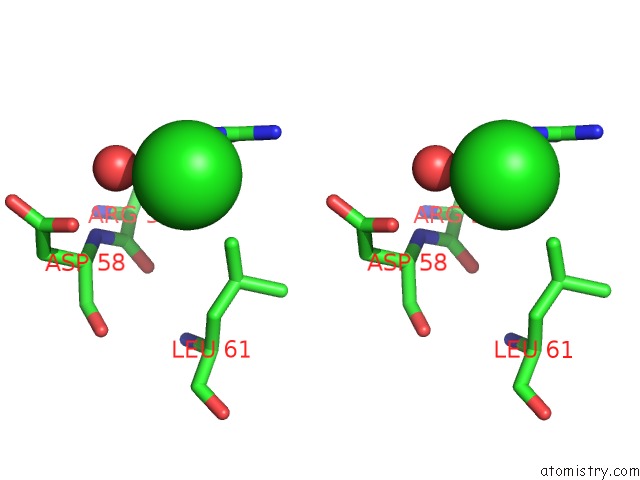
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of X-Ray Structure Dps From Kineococcus Radiotolerans in Complex with Mn (II) Ions. within 5.0Å range:
|
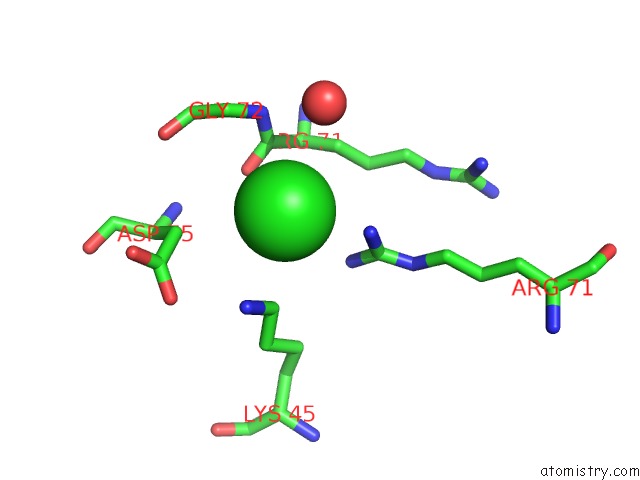
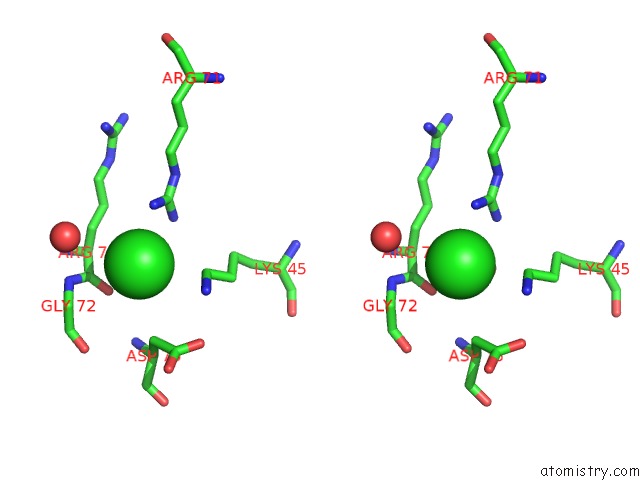
Chlorine binding site 3 out of 6 in 4a25
Go back to
Chlorine binding site 3 out
of 6 in the X-Ray Structure Dps From Kineococcus Radiotolerans in Complex with Mn (II) Ions.
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of X-Ray Structure Dps From Kineococcus Radiotolerans in Complex with Mn (II) Ions. within 5.0Å range:
|
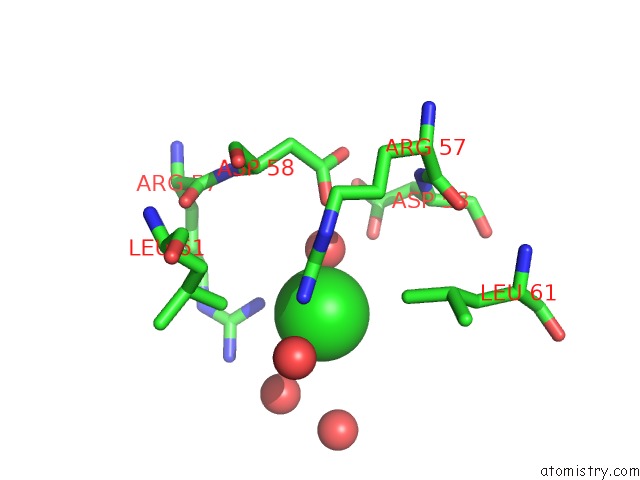
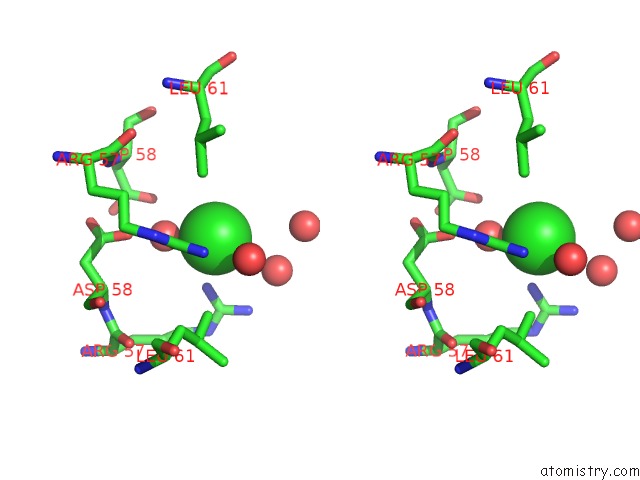
Chlorine binding site 4 out of 6 in 4a25
Go back to
Chlorine binding site 4 out
of 6 in the X-Ray Structure Dps From Kineococcus Radiotolerans in Complex with Mn (II) Ions.
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of X-Ray Structure Dps From Kineococcus Radiotolerans in Complex with Mn (II) Ions. within 5.0Å range:
|
Chlorine binding site 5 out of 6 in 4a25
Go back to
Chlorine binding site 5 out
of 6 in the X-Ray Structure Dps From Kineococcus Radiotolerans in Complex with Mn (II) Ions.
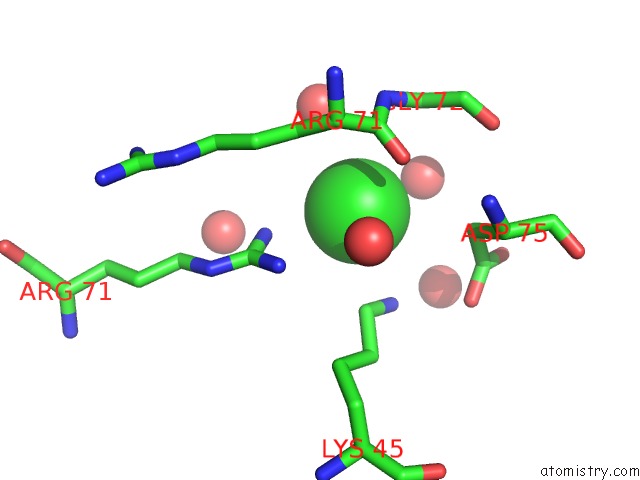
Mono view
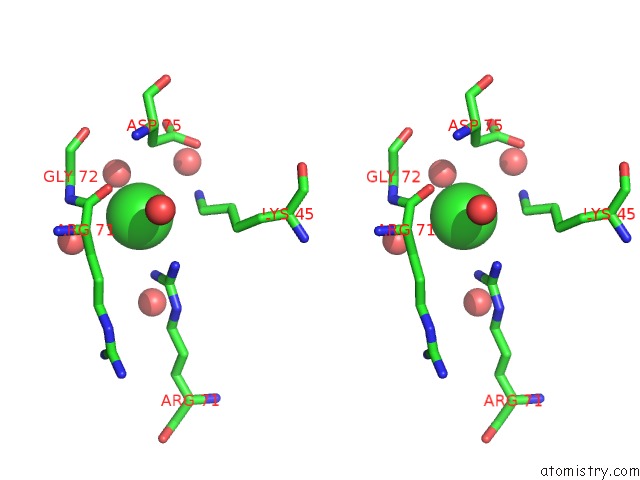
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 5 of X-Ray Structure Dps From Kineococcus Radiotolerans in Complex with Mn (II) Ions. within 5.0Å range:
|
Chlorine binding site 6 out of 6 in 4a25
Go back to
Chlorine binding site 6 out
of 6 in the X-Ray Structure Dps From Kineococcus Radiotolerans in Complex with Mn (II) Ions.
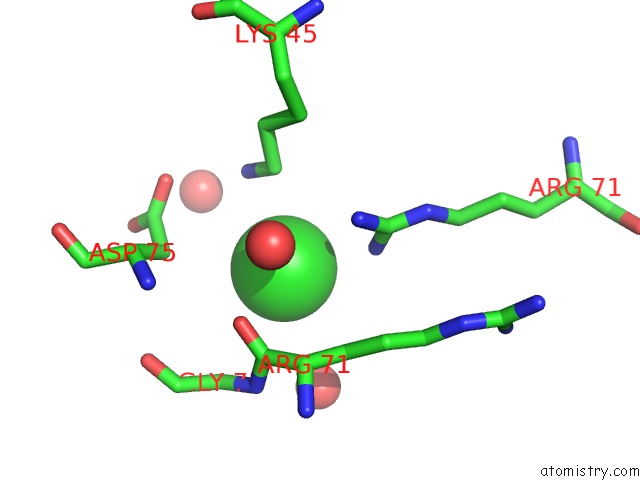
Mono view
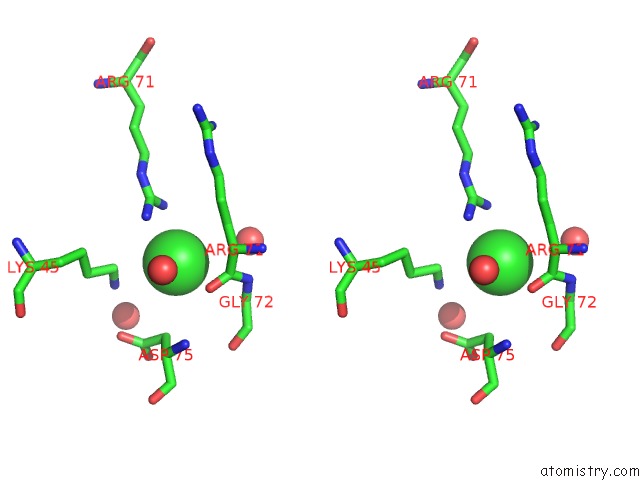
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 6 of X-Ray Structure Dps From Kineococcus Radiotolerans in Complex with Mn (II) Ions. within 5.0Å range:
|
Reference:
M.Ardini,
A.Fiorillo,
M.Fittipaldi,
S.Stefanini,
D.Gatteschi,
A.Llari,
E.Chiancone.
Kineococcus Radiotolerans Dps Forms A Heteronuclear Mn-Fe Ferroxidase Center That May Explain the Mn-Dependent Protection Against Oxidative Stress. Biochim.Biophys.Acta V.1830 3745 2013.
ISSN: ISSN 0006-3002
PubMed: 23396000
DOI: 10.1016/J.BBAGEN.2013.02.003
Page generated: Fri Jul 11 12:42:28 2025
ISSN: ISSN 0006-3002
PubMed: 23396000
DOI: 10.1016/J.BBAGEN.2013.02.003
Last articles
K in 7JUJK in 7JVU
K in 7JOM
K in 7JKU
K in 7JPS
K in 7JPO
K in 7JMV
K in 7JMR
K in 7JFN
K in 7G7Y