Chlorine »
PDB 4fv3-4g7y »
4fz4 »
Chlorine in PDB 4fz4: Crystal Structure of HP0197-18KD
Protein crystallography data
The structure of Crystal Structure of HP0197-18KD, PDB code: 4fz4
was solved by
Z.Yuan,
X.Yan,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 18.82 / 2.44 |
| Space group | P 21 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 49.235, 87.601, 38.540, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 22.2 / 26.3 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Crystal Structure of HP0197-18KD
(pdb code 4fz4). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 2 binding sites of Chlorine where determined in the Crystal Structure of HP0197-18KD, PDB code: 4fz4:
Jump to Chlorine binding site number: 1; 2;
In total 2 binding sites of Chlorine where determined in the Crystal Structure of HP0197-18KD, PDB code: 4fz4:
Jump to Chlorine binding site number: 1; 2;
Chlorine binding site 1 out of 2 in 4fz4
Go back to
Chlorine binding site 1 out
of 2 in the Crystal Structure of HP0197-18KD
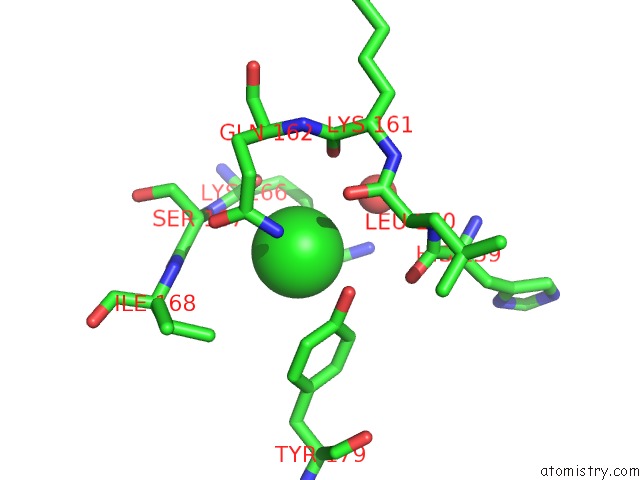
Mono view
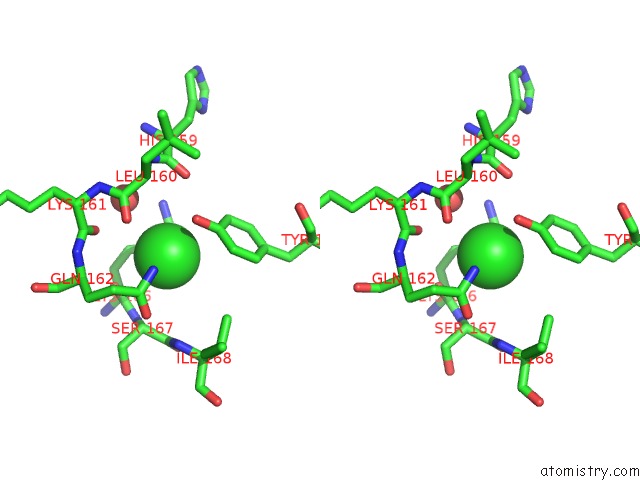
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Crystal Structure of HP0197-18KD within 5.0Å range:
|
Chlorine binding site 2 out of 2 in 4fz4
Go back to
Chlorine binding site 2 out
of 2 in the Crystal Structure of HP0197-18KD
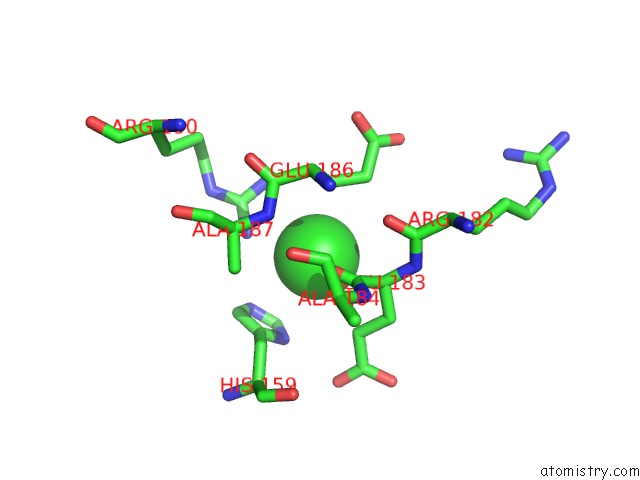
Mono view
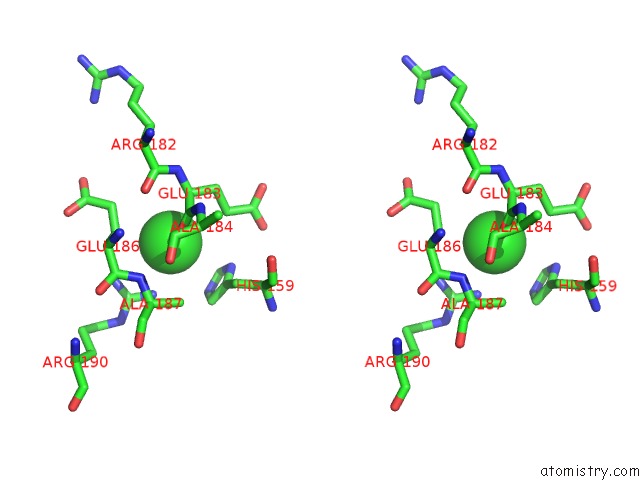
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Crystal Structure of HP0197-18KD within 5.0Å range:
|
Reference:
Z.Z.Yuan,
X.J.Yan,
A.D.Zhang,
B.Chen,
Y.Q.Shen,
M.L.Jin.
Molecular Mechanism By Which Surface Antigen HP0197 Mediates Host Cell Attachment in the Pathogenic Bacteria Streptococcus Suis J.Biol.Chem. V. 288 956 2013.
ISSN: ISSN 0021-9258
PubMed: 23184929
DOI: 10.1074/JBC.M112.388686
Page generated: Fri Jul 11 15:26:49 2025
ISSN: ISSN 0021-9258
PubMed: 23184929
DOI: 10.1074/JBC.M112.388686
Last articles
Mg in 4GMJMg in 4GNK
Mg in 4GNI
Mg in 4GN0
Mg in 4GMX
Mg in 4GME
Mg in 4GKM
Mg in 4GKR
Mg in 4GHL
Mg in 4GIU