Chlorine »
PDB 4g81-4gen »
4g9s »
Chlorine in PDB 4g9s: Crystal Structure of Escherichia Coli Plig in Complex with Atlantic Salmon G-Type Lysozyme
Protein crystallography data
The structure of Crystal Structure of Escherichia Coli Plig in Complex with Atlantic Salmon G-Type Lysozyme, PDB code: 4g9s
was solved by
S.Leysen,
L.Vanderkelen,
S.D.Weeks,
C.W.Michiels,
S.V.Strelkov,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 43.27 / 0.95 |
| Space group | P 65 |
| Cell size a, b, c (Å), α, β, γ (°) | 132.182, 132.182, 42.902, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 12.9 / 13.6 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Crystal Structure of Escherichia Coli Plig in Complex with Atlantic Salmon G-Type Lysozyme
(pdb code 4g9s). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total only one binding site of Chlorine was determined in the Crystal Structure of Escherichia Coli Plig in Complex with Atlantic Salmon G-Type Lysozyme, PDB code: 4g9s:
In total only one binding site of Chlorine was determined in the Crystal Structure of Escherichia Coli Plig in Complex with Atlantic Salmon G-Type Lysozyme, PDB code: 4g9s:
Chlorine binding site 1 out of 1 in 4g9s
Go back to
Chlorine binding site 1 out
of 1 in the Crystal Structure of Escherichia Coli Plig in Complex with Atlantic Salmon G-Type Lysozyme
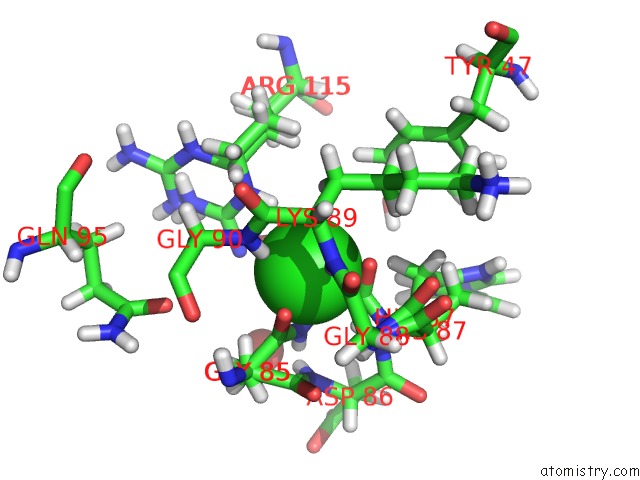
Mono view
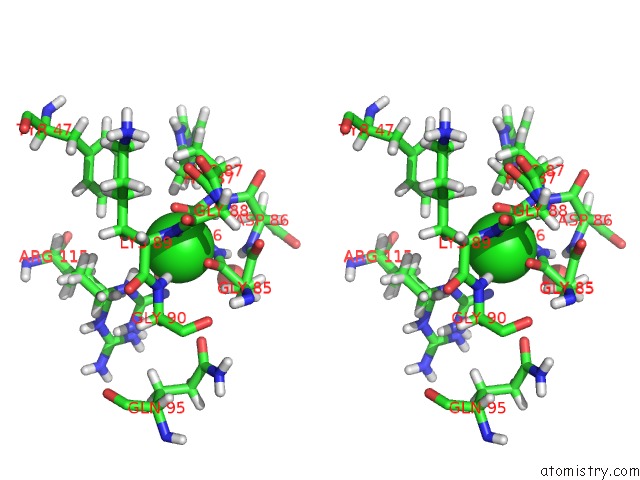
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Crystal Structure of Escherichia Coli Plig in Complex with Atlantic Salmon G-Type Lysozyme within 5.0Å range:
|
Reference:
S.Leysen,
L.Vanderkelen,
S.D.Weeks,
C.W.Michiels,
S.V.Strelkov.
Structural Basis of Bacterial Defense Against G-Type Lysozyme-Based Innate Immunity. Cell.Mol.Life Sci. V. 70 1113 2013.
ISSN: ISSN 1420-682X
PubMed: 23086131
DOI: 10.1007/S00018-012-1184-1
Page generated: Fri Jul 11 15:34:03 2025
ISSN: ISSN 1420-682X
PubMed: 23086131
DOI: 10.1007/S00018-012-1184-1
Last articles
Mg in 6CFRMg in 6CFO
Mg in 6CER
Mg in 6CC3
Mg in 6CEO
Mg in 6CDW
Mg in 6CCX
Mg in 6CCK
Mg in 6CCH
Mg in 6CCE