Chlorine »
PDB 4g81-4gen »
4gcc »
Chlorine in PDB 4gcc: Room Temperature X-Ray Diffraction Study of A 6-Fold Molar Excess of A Cisplatin/Carboplatin Mixture Binding to Hewl, Dataset 1
Enzymatic activity of Room Temperature X-Ray Diffraction Study of A 6-Fold Molar Excess of A Cisplatin/Carboplatin Mixture Binding to Hewl, Dataset 1
All present enzymatic activity of Room Temperature X-Ray Diffraction Study of A 6-Fold Molar Excess of A Cisplatin/Carboplatin Mixture Binding to Hewl, Dataset 1:
3.2.1.17;
3.2.1.17;
Protein crystallography data
The structure of Room Temperature X-Ray Diffraction Study of A 6-Fold Molar Excess of A Cisplatin/Carboplatin Mixture Binding to Hewl, Dataset 1, PDB code: 4gcc
was solved by
J.R.Helliwell,
S.W.M.Tanley,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 56.47 / 2.00 |
| Space group | P 43 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 79.863, 79.863, 37.802, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 18.6 / 24.8 |
Other elements in 4gcc:
The structure of Room Temperature X-Ray Diffraction Study of A 6-Fold Molar Excess of A Cisplatin/Carboplatin Mixture Binding to Hewl, Dataset 1 also contains other interesting chemical elements:
| Platinum | (Pt) | 2 atoms |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Room Temperature X-Ray Diffraction Study of A 6-Fold Molar Excess of A Cisplatin/Carboplatin Mixture Binding to Hewl, Dataset 1
(pdb code 4gcc). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 2 binding sites of Chlorine where determined in the Room Temperature X-Ray Diffraction Study of A 6-Fold Molar Excess of A Cisplatin/Carboplatin Mixture Binding to Hewl, Dataset 1, PDB code: 4gcc:
Jump to Chlorine binding site number: 1; 2;
In total 2 binding sites of Chlorine where determined in the Room Temperature X-Ray Diffraction Study of A 6-Fold Molar Excess of A Cisplatin/Carboplatin Mixture Binding to Hewl, Dataset 1, PDB code: 4gcc:
Jump to Chlorine binding site number: 1; 2;
Chlorine binding site 1 out of 2 in 4gcc
Go back to
Chlorine binding site 1 out
of 2 in the Room Temperature X-Ray Diffraction Study of A 6-Fold Molar Excess of A Cisplatin/Carboplatin Mixture Binding to Hewl, Dataset 1
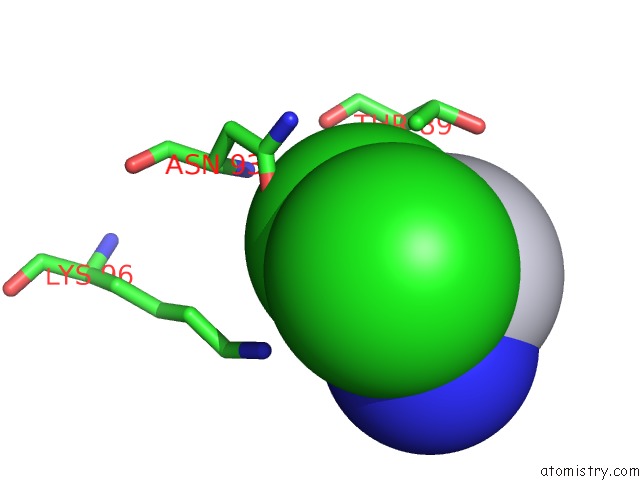
Mono view
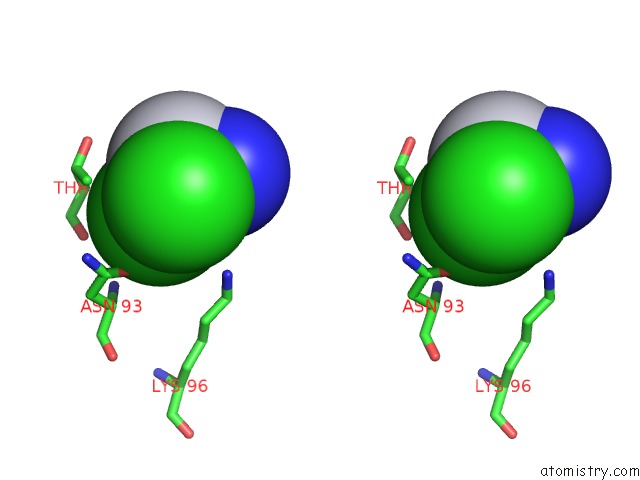
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Room Temperature X-Ray Diffraction Study of A 6-Fold Molar Excess of A Cisplatin/Carboplatin Mixture Binding to Hewl, Dataset 1 within 5.0Å range:
|
Chlorine binding site 2 out of 2 in 4gcc
Go back to
Chlorine binding site 2 out
of 2 in the Room Temperature X-Ray Diffraction Study of A 6-Fold Molar Excess of A Cisplatin/Carboplatin Mixture Binding to Hewl, Dataset 1
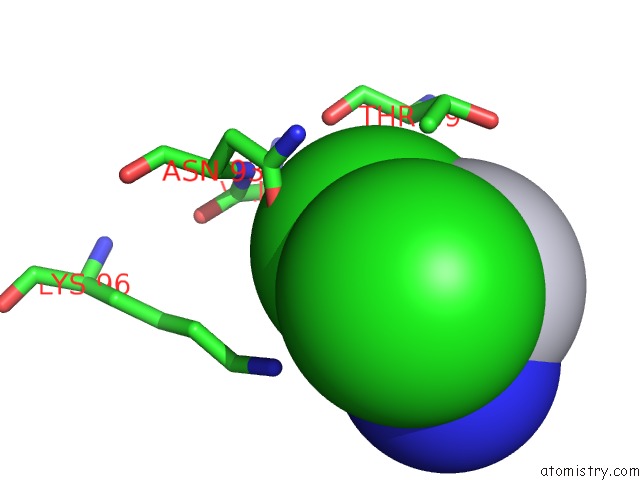
Mono view
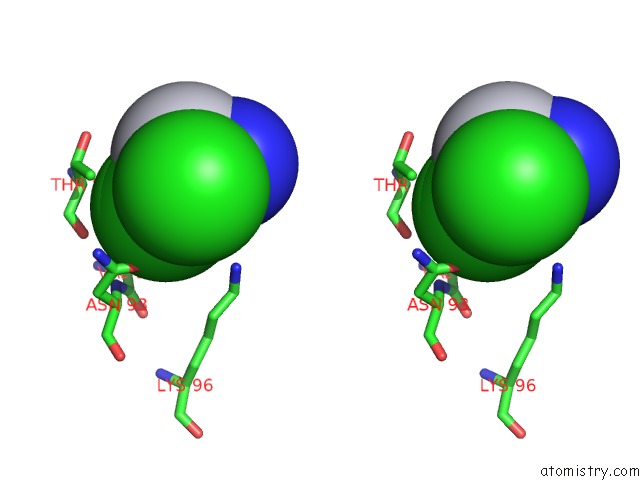
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Room Temperature X-Ray Diffraction Study of A 6-Fold Molar Excess of A Cisplatin/Carboplatin Mixture Binding to Hewl, Dataset 1 within 5.0Å range:
|
Reference:
J.R.Helliwell,
S.W.Tanley.
The Crystal Structure Analysis of the Relative Binding of Cisplatin and Carboplatin in A Mixture with Histidine in A Protein Studied at 100 and 300 K with Repeated X-Ray Irradiation. Acta Crystallogr.,Sect.D V. 69 121 2013.
ISSN: ISSN 0907-4449
PubMed: 23275170
DOI: 10.1107/S090744491204423X
Page generated: Fri Jul 11 15:35:43 2025
ISSN: ISSN 0907-4449
PubMed: 23275170
DOI: 10.1107/S090744491204423X
Last articles
Mg in 6IK9Mg in 6IJI
Mg in 6IJH
Mg in 6IID
Mg in 6III
Mg in 6IHW
Mg in 6II6
Mg in 6IHV
Mg in 6IHU
Mg in 6IHS