Chlorine »
PDB 4gta-4h1t »
4guf »
Chlorine in PDB 4guf: 1.5 Angstrom Crystal Structure of the Salmonella Enterica 3- Dehydroquinate Dehydratase (Arod) E86A Mutant
Enzymatic activity of 1.5 Angstrom Crystal Structure of the Salmonella Enterica 3- Dehydroquinate Dehydratase (Arod) E86A Mutant
All present enzymatic activity of 1.5 Angstrom Crystal Structure of the Salmonella Enterica 3- Dehydroquinate Dehydratase (Arod) E86A Mutant:
4.2.1.10;
4.2.1.10;
Protein crystallography data
The structure of 1.5 Angstrom Crystal Structure of the Salmonella Enterica 3- Dehydroquinate Dehydratase (Arod) E86A Mutant, PDB code: 4guf
was solved by
S.H.Light,
G.Minasov,
M.-E.Duban,
L.Shuvalova,
K.Kwon,
A.Lavie,
W.F.Anderson,
Center For Structural Genomics Of Infectious Diseases(Csgid),
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 29.43 / 1.50 |
| Space group | P 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 36.771, 45.552, 81.059, 93.90, 101.41, 105.90 |
| R / Rfree (%) | 16.7 / 19.1 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the 1.5 Angstrom Crystal Structure of the Salmonella Enterica 3- Dehydroquinate Dehydratase (Arod) E86A Mutant
(pdb code 4guf). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total only one binding site of Chlorine was determined in the 1.5 Angstrom Crystal Structure of the Salmonella Enterica 3- Dehydroquinate Dehydratase (Arod) E86A Mutant, PDB code: 4guf:
In total only one binding site of Chlorine was determined in the 1.5 Angstrom Crystal Structure of the Salmonella Enterica 3- Dehydroquinate Dehydratase (Arod) E86A Mutant, PDB code: 4guf:
Chlorine binding site 1 out of 1 in 4guf
Go back to
Chlorine binding site 1 out
of 1 in the 1.5 Angstrom Crystal Structure of the Salmonella Enterica 3- Dehydroquinate Dehydratase (Arod) E86A Mutant
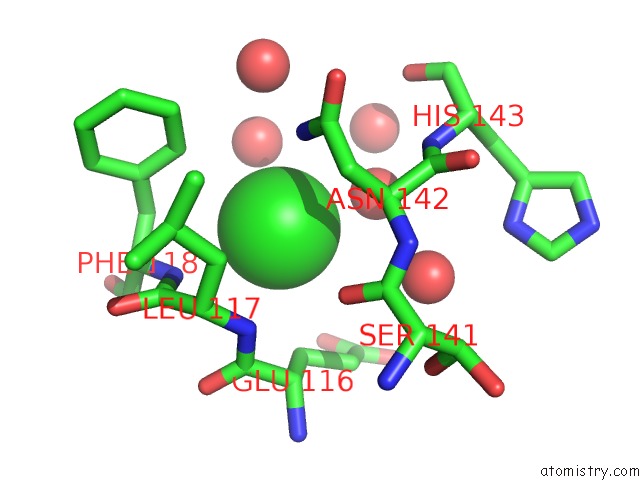
Mono view
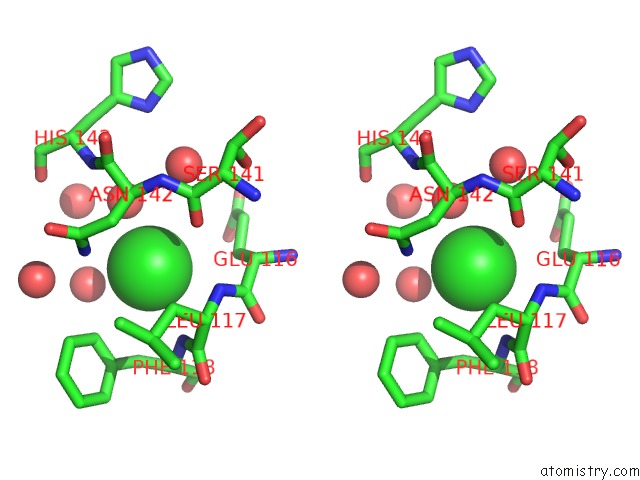
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of 1.5 Angstrom Crystal Structure of the Salmonella Enterica 3- Dehydroquinate Dehydratase (Arod) E86A Mutant within 5.0Å range:
|
Reference:
S.H.Light,
W.F.Anderson,
A.Lavie.
Reassessing the Type I Dehydroquinate Dehydratase Catalytic Triad: Kinetic and Structural Studies of GLU86 Mutants. Protein Sci. V. 22 418 2013.
ISSN: ISSN 0961-8368
PubMed: 23341204
DOI: 10.1002/PRO.2218
Page generated: Fri Jul 11 15:55:25 2025
ISSN: ISSN 0961-8368
PubMed: 23341204
DOI: 10.1002/PRO.2218
Last articles
Mg in 6PRVMg in 6PSS
Mg in 6PSR
Mg in 6PSQ
Mg in 6PRY
Mg in 6PRU
Mg in 6PRC
Mg in 6PR5
Mg in 6PQV
Mg in 6PQR