Chlorine »
PDB 4j93-4jk9 »
4jh0 »
Chlorine in PDB 4jh0: Crystal Structure of Dipeptidyl-Peptidase 4 (CD26, Adenosine Deaminase Complexing Protein 2) (Dpp-IV-Wt) Complex with Bms-767778 Aka 2-(3- (Aminomethyl)-4-(2,4- Dichlorophenyl)-2-Methyl-5-Oxo-5,7-Dihydro-6H- Pyrrolo[3,4- B]Pyridin-6-Yl)-N,N-Dimethylacetamide
Enzymatic activity of Crystal Structure of Dipeptidyl-Peptidase 4 (CD26, Adenosine Deaminase Complexing Protein 2) (Dpp-IV-Wt) Complex with Bms-767778 Aka 2-(3- (Aminomethyl)-4-(2,4- Dichlorophenyl)-2-Methyl-5-Oxo-5,7-Dihydro-6H- Pyrrolo[3,4- B]Pyridin-6-Yl)-N,N-Dimethylacetamide
All present enzymatic activity of Crystal Structure of Dipeptidyl-Peptidase 4 (CD26, Adenosine Deaminase Complexing Protein 2) (Dpp-IV-Wt) Complex with Bms-767778 Aka 2-(3- (Aminomethyl)-4-(2,4- Dichlorophenyl)-2-Methyl-5-Oxo-5,7-Dihydro-6H- Pyrrolo[3,4- B]Pyridin-6-Yl)-N,N-Dimethylacetamide:
3.4.14.5;
3.4.14.5;
Protein crystallography data
The structure of Crystal Structure of Dipeptidyl-Peptidase 4 (CD26, Adenosine Deaminase Complexing Protein 2) (Dpp-IV-Wt) Complex with Bms-767778 Aka 2-(3- (Aminomethyl)-4-(2,4- Dichlorophenyl)-2-Methyl-5-Oxo-5,7-Dihydro-6H- Pyrrolo[3,4- B]Pyridin-6-Yl)-N,N-Dimethylacetamide, PDB code: 4jh0
was solved by
H.E.Klei,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 47.95 / 2.35 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 65.311, 67.105, 423.771, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 24.3 / 27.2 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Crystal Structure of Dipeptidyl-Peptidase 4 (CD26, Adenosine Deaminase Complexing Protein 2) (Dpp-IV-Wt) Complex with Bms-767778 Aka 2-(3- (Aminomethyl)-4-(2,4- Dichlorophenyl)-2-Methyl-5-Oxo-5,7-Dihydro-6H- Pyrrolo[3,4- B]Pyridin-6-Yl)-N,N-Dimethylacetamide
(pdb code 4jh0). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 4 binding sites of Chlorine where determined in the Crystal Structure of Dipeptidyl-Peptidase 4 (CD26, Adenosine Deaminase Complexing Protein 2) (Dpp-IV-Wt) Complex with Bms-767778 Aka 2-(3- (Aminomethyl)-4-(2,4- Dichlorophenyl)-2-Methyl-5-Oxo-5,7-Dihydro-6H- Pyrrolo[3,4- B]Pyridin-6-Yl)-N,N-Dimethylacetamide, PDB code: 4jh0:
Jump to Chlorine binding site number: 1; 2; 3; 4;
In total 4 binding sites of Chlorine where determined in the Crystal Structure of Dipeptidyl-Peptidase 4 (CD26, Adenosine Deaminase Complexing Protein 2) (Dpp-IV-Wt) Complex with Bms-767778 Aka 2-(3- (Aminomethyl)-4-(2,4- Dichlorophenyl)-2-Methyl-5-Oxo-5,7-Dihydro-6H- Pyrrolo[3,4- B]Pyridin-6-Yl)-N,N-Dimethylacetamide, PDB code: 4jh0:
Jump to Chlorine binding site number: 1; 2; 3; 4;
Chlorine binding site 1 out of 4 in 4jh0
Go back to
Chlorine binding site 1 out
of 4 in the Crystal Structure of Dipeptidyl-Peptidase 4 (CD26, Adenosine Deaminase Complexing Protein 2) (Dpp-IV-Wt) Complex with Bms-767778 Aka 2-(3- (Aminomethyl)-4-(2,4- Dichlorophenyl)-2-Methyl-5-Oxo-5,7-Dihydro-6H- Pyrrolo[3,4- B]Pyridin-6-Yl)-N,N-Dimethylacetamide
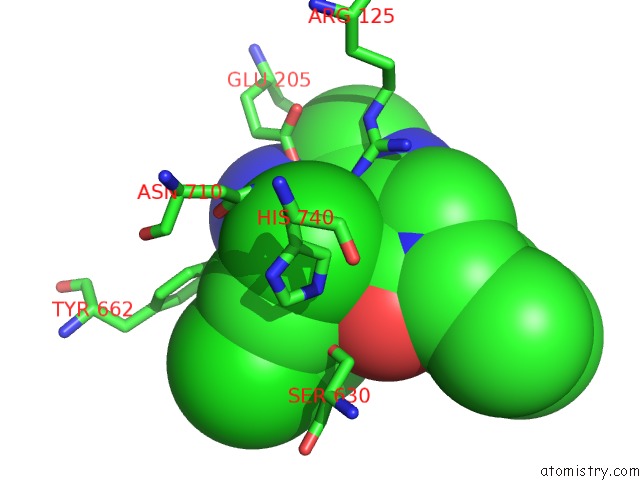
Mono view
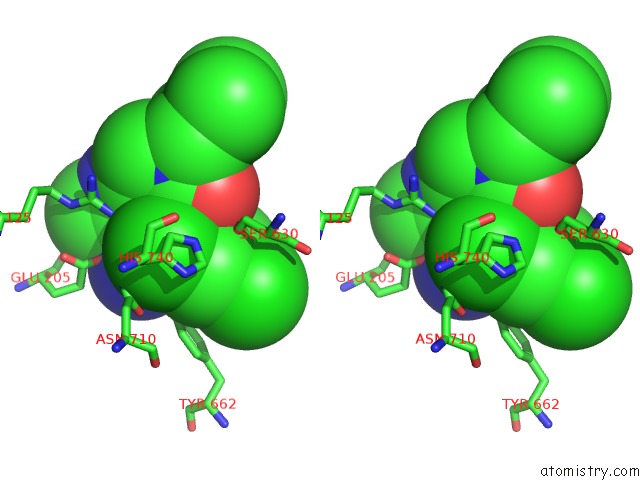
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Crystal Structure of Dipeptidyl-Peptidase 4 (CD26, Adenosine Deaminase Complexing Protein 2) (Dpp-IV-Wt) Complex with Bms-767778 Aka 2-(3- (Aminomethyl)-4-(2,4- Dichlorophenyl)-2-Methyl-5-Oxo-5,7-Dihydro-6H- Pyrrolo[3,4- B]Pyridin-6-Yl)-N,N-Dimethylacetamide within 5.0Å range:
|
Chlorine binding site 2 out of 4 in 4jh0
Go back to
Chlorine binding site 2 out
of 4 in the Crystal Structure of Dipeptidyl-Peptidase 4 (CD26, Adenosine Deaminase Complexing Protein 2) (Dpp-IV-Wt) Complex with Bms-767778 Aka 2-(3- (Aminomethyl)-4-(2,4- Dichlorophenyl)-2-Methyl-5-Oxo-5,7-Dihydro-6H- Pyrrolo[3,4- B]Pyridin-6-Yl)-N,N-Dimethylacetamide
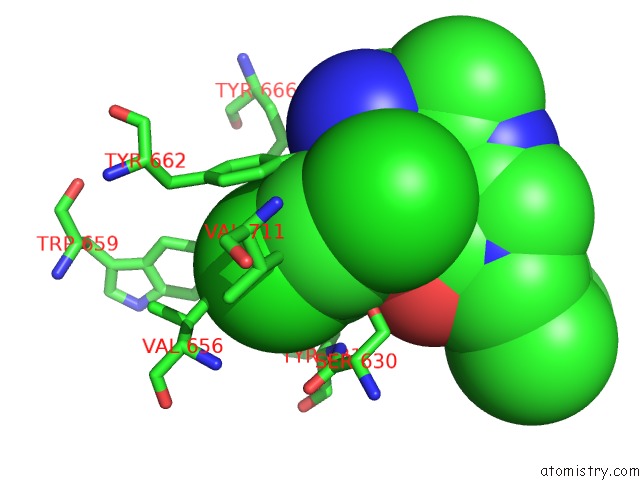
Mono view
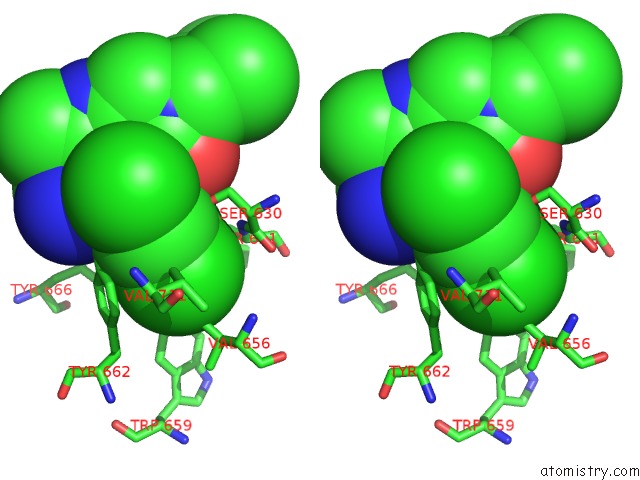
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Crystal Structure of Dipeptidyl-Peptidase 4 (CD26, Adenosine Deaminase Complexing Protein 2) (Dpp-IV-Wt) Complex with Bms-767778 Aka 2-(3- (Aminomethyl)-4-(2,4- Dichlorophenyl)-2-Methyl-5-Oxo-5,7-Dihydro-6H- Pyrrolo[3,4- B]Pyridin-6-Yl)-N,N-Dimethylacetamide within 5.0Å range:
|
Chlorine binding site 3 out of 4 in 4jh0
Go back to
Chlorine binding site 3 out
of 4 in the Crystal Structure of Dipeptidyl-Peptidase 4 (CD26, Adenosine Deaminase Complexing Protein 2) (Dpp-IV-Wt) Complex with Bms-767778 Aka 2-(3- (Aminomethyl)-4-(2,4- Dichlorophenyl)-2-Methyl-5-Oxo-5,7-Dihydro-6H- Pyrrolo[3,4- B]Pyridin-6-Yl)-N,N-Dimethylacetamide
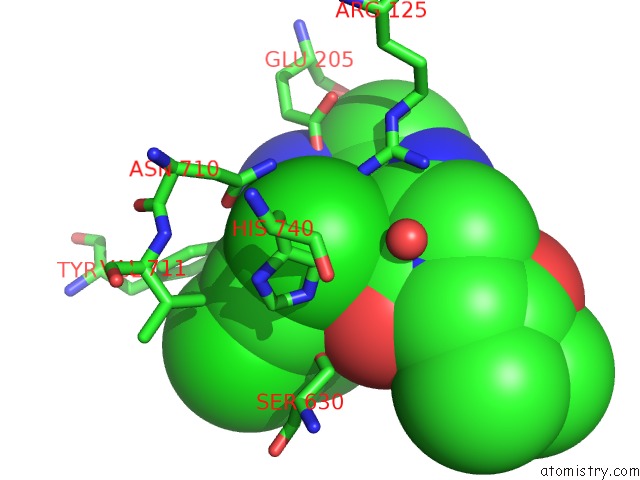
Mono view
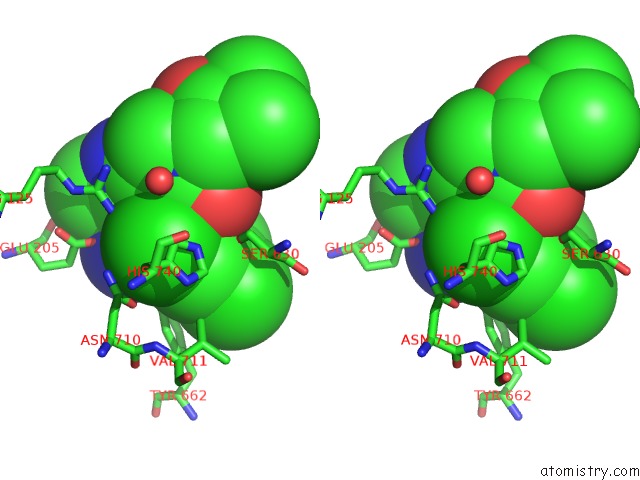
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Crystal Structure of Dipeptidyl-Peptidase 4 (CD26, Adenosine Deaminase Complexing Protein 2) (Dpp-IV-Wt) Complex with Bms-767778 Aka 2-(3- (Aminomethyl)-4-(2,4- Dichlorophenyl)-2-Methyl-5-Oxo-5,7-Dihydro-6H- Pyrrolo[3,4- B]Pyridin-6-Yl)-N,N-Dimethylacetamide within 5.0Å range:
|
Chlorine binding site 4 out of 4 in 4jh0
Go back to
Chlorine binding site 4 out
of 4 in the Crystal Structure of Dipeptidyl-Peptidase 4 (CD26, Adenosine Deaminase Complexing Protein 2) (Dpp-IV-Wt) Complex with Bms-767778 Aka 2-(3- (Aminomethyl)-4-(2,4- Dichlorophenyl)-2-Methyl-5-Oxo-5,7-Dihydro-6H- Pyrrolo[3,4- B]Pyridin-6-Yl)-N,N-Dimethylacetamide
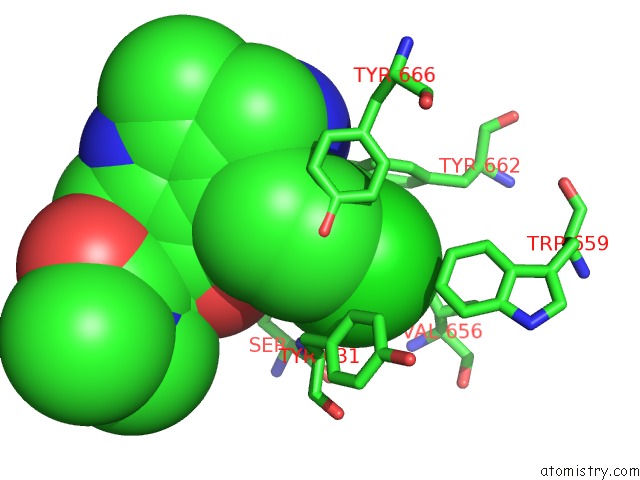
Mono view
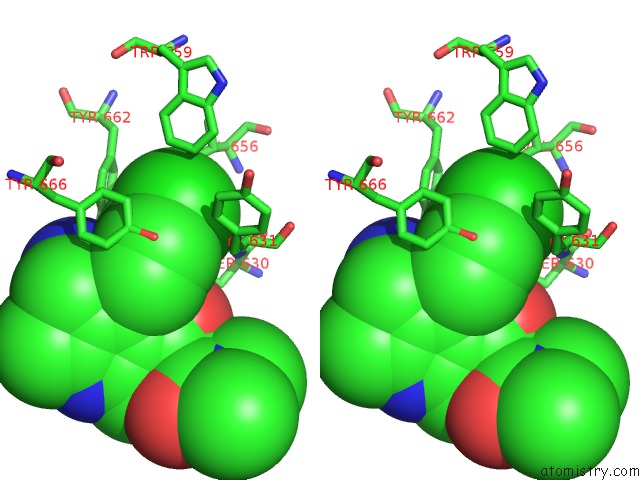
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of Crystal Structure of Dipeptidyl-Peptidase 4 (CD26, Adenosine Deaminase Complexing Protein 2) (Dpp-IV-Wt) Complex with Bms-767778 Aka 2-(3- (Aminomethyl)-4-(2,4- Dichlorophenyl)-2-Methyl-5-Oxo-5,7-Dihydro-6H- Pyrrolo[3,4- B]Pyridin-6-Yl)-N,N-Dimethylacetamide within 5.0Å range:
|
Reference:
P.Devasthale,
Y.Wang,
W.Wang,
J.Fevig,
J.Feng,
A.Wang,
T.Harrity,
D.Egan,
N.Morgan,
M.Cap,
A.Fura,
H.E.Klei,
K.Kish,
C.Weigelt,
L.Sun,
P.Levesque,
F.Moulin,
Y.X.Li,
R.Zahler,
M.S.Kirby,
L.G.Hamann.
Optimization of Activity, Selectivity, and Liability Profiles in 5-Oxopyrrolopyridine DPP4 Inhibitors Leading to Clinical Candidate (Sa)-2-(3-(Aminomethyl)-4-(2,4-Dichlorophenyl)-2-Methyl-5- Oxo-5H-Pyrrolo[3,4-B]Pyridin-6(7H)-Yl)-N,N-Dimethylacetamide (Bms-767778). J.Med.Chem. V. 56 7343 2013.
ISSN: ISSN 0022-2623
PubMed: 23964740
DOI: 10.1021/JM4008906
Page generated: Fri Jul 11 17:19:25 2025
ISSN: ISSN 0022-2623
PubMed: 23964740
DOI: 10.1021/JM4008906
Last articles
Mg in 5C4JMg in 5C4M
Mg in 5C4I
Mg in 5C4A
Mg in 5C2G
Mg in 5C4G
Mg in 5C45
Mg in 5C3E
Mg in 5C46
Mg in 5C2W