Chlorine »
PDB 4me9-4ml5 »
4mf3 »
Chlorine in PDB 4mf3: Crystal Structure of Human GRIK1 Complexed with A 6-(Tetrazolyl)Aryl Decahydroisoquinoline Antagonist
Protein crystallography data
The structure of Crystal Structure of Human GRIK1 Complexed with A 6-(Tetrazolyl)Aryl Decahydroisoquinoline Antagonist, PDB code: 4mf3
was solved by
J.A.Martinez-Perez,
S.Iyengar,
H.E.Shannon,
D.Bleakman,
A.Alt,
D.K.Clawson,
B.M.Arnold,
M.G.Bell,
T.J.Bleisch,
A.M.Castano,
M.Del Prado,
E.Dominguez,
A.M.Escribano,
S.A.Filla,
K.H.Ho,
K.J.Hudziak,
C.K.Jones,
M.A.Katofiasc,
A.Mateo,
B.M.Mathes,
E.L.Mattiuz,
A.M.L.Ogden,
L.A.Phebus,
R.M.A.Simmons,
D.R.Stack,
R.E.Stratford,
M.A.Winter,
Z.Wu,
P.L.Ornstein,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 20.00 / 3.00 |
| Space group | P 32 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 108.018, 108.018, 109.766, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 23.7 / 26 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Crystal Structure of Human GRIK1 Complexed with A 6-(Tetrazolyl)Aryl Decahydroisoquinoline Antagonist
(pdb code 4mf3). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 2 binding sites of Chlorine where determined in the Crystal Structure of Human GRIK1 Complexed with A 6-(Tetrazolyl)Aryl Decahydroisoquinoline Antagonist, PDB code: 4mf3:
Jump to Chlorine binding site number: 1; 2;
In total 2 binding sites of Chlorine where determined in the Crystal Structure of Human GRIK1 Complexed with A 6-(Tetrazolyl)Aryl Decahydroisoquinoline Antagonist, PDB code: 4mf3:
Jump to Chlorine binding site number: 1; 2;
Chlorine binding site 1 out of 2 in 4mf3
Go back to
Chlorine binding site 1 out
of 2 in the Crystal Structure of Human GRIK1 Complexed with A 6-(Tetrazolyl)Aryl Decahydroisoquinoline Antagonist
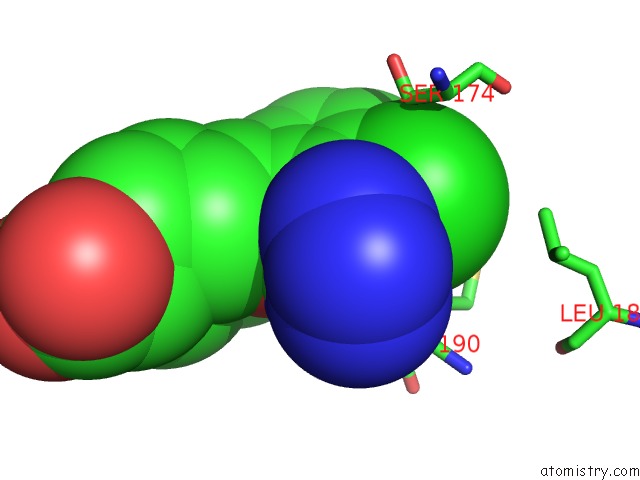
Mono view
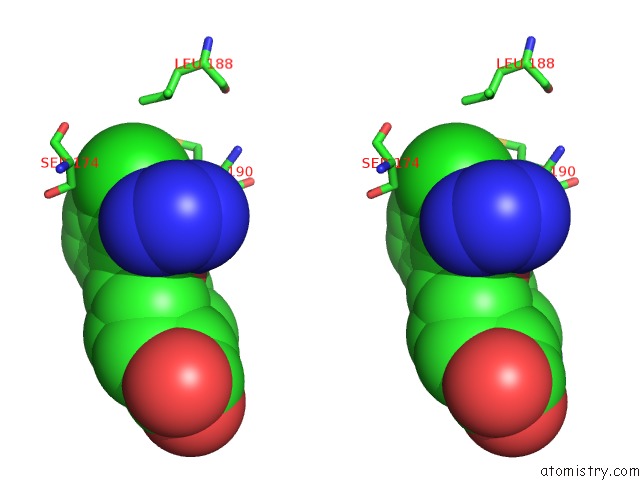
Stereo pair view

Mono view

Stereo pair view
|
|
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Crystal Structure of Human GRIK1 Complexed with A 6-(Tetrazolyl)Aryl Decahydroisoquinoline Antagonist within 5.0Å range:
|
Chlorine binding site 2 out of 2 in 4mf3
Go back to
Chlorine binding site 2 out
of 2 in the Crystal Structure of Human GRIK1 Complexed with A 6-(Tetrazolyl)Aryl Decahydroisoquinoline Antagonist
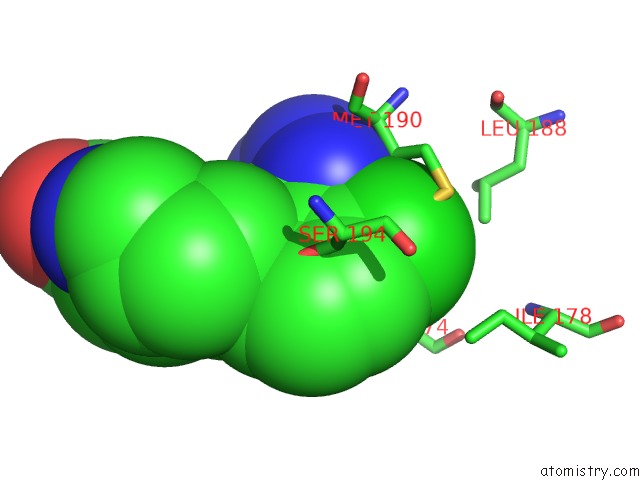
Mono view
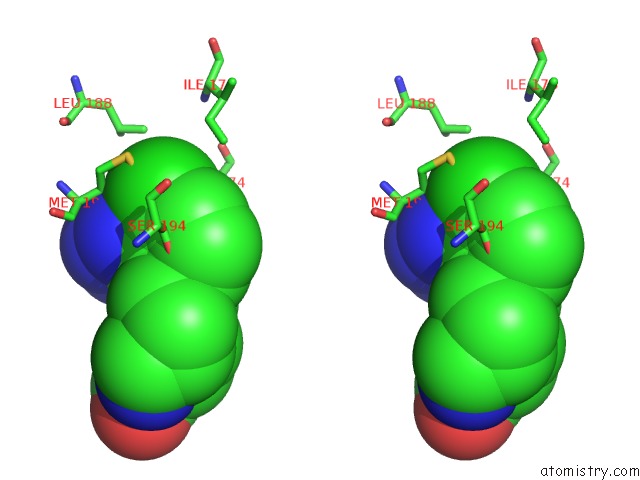
Stereo pair view

Mono view

Stereo pair view
|
|
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Crystal Structure of Human GRIK1 Complexed with A 6-(Tetrazolyl)Aryl Decahydroisoquinoline Antagonist within 5.0Å range:
|
Reference:
J.A.Martinez-Perez,
S.Iyengar,
H.E.Shannon,
D.Bleakman,
A.Alt,
D.K.Clawson,
B.M.Arnold,
M.G.Bell,
T.J.Bleisch,
A.M.Castano,
M.Del Prado,
E.Dominguez,
A.M.Escribano,
S.A.Filla,
K.H.Ho,
K.J.Hudziak,
C.K.Jones,
A.Mateo,
B.M.Mathes,
E.L.Mattiuz,
A.M.Ogden,
R.M.Simmons,
D.R.Stack,
R.E.Stratford,
M.A.Winter,
Z.Wu,
P.L.Ornstein.
GLUK1 Antagonists From 6-(Tetrazolyl)Phenyl Decahydroisoquinoline Derivatives: in Vitro Profile and in Vivo Analgesic Efficacy. Bioorg.Med.Chem.Lett. V. 23 6463 2013.
ISSN: ISSN 0960-894X
PubMed: 24140446
DOI: 10.1016/J.BMCL.2013.09.045
Page generated: Fri Jul 11 19:03:10 2025
ISSN: ISSN 0960-894X
PubMed: 24140446
DOI: 10.1016/J.BMCL.2013.09.045
Last articles
W in 1DV4W in 1FR3
W in 1GUG
W in 1H9R
W in 1H9K
W in 1H0H
W in 1FEZ
W in 1FKA
W in 1E3P
W in 1E18