Chlorine »
PDB 4q69-4qgp »
4qba »
Chlorine in PDB 4qba: Crystal Structure of the Effector-Binding Domain of S. Aureus Ccpe
Protein crystallography data
The structure of Crystal Structure of the Effector-Binding Domain of S. Aureus Ccpe, PDB code: 4qba
was solved by
X.Liu,
L.Lan,
C.G.Yang,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 30.00 / 2.21 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 73.605, 77.182, 93.968, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 20.2 / 24.3 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Crystal Structure of the Effector-Binding Domain of S. Aureus Ccpe
(pdb code 4qba). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 2 binding sites of Chlorine where determined in the Crystal Structure of the Effector-Binding Domain of S. Aureus Ccpe, PDB code: 4qba:
Jump to Chlorine binding site number: 1; 2;
In total 2 binding sites of Chlorine where determined in the Crystal Structure of the Effector-Binding Domain of S. Aureus Ccpe, PDB code: 4qba:
Jump to Chlorine binding site number: 1; 2;
Chlorine binding site 1 out of 2 in 4qba
Go back to
Chlorine binding site 1 out
of 2 in the Crystal Structure of the Effector-Binding Domain of S. Aureus Ccpe
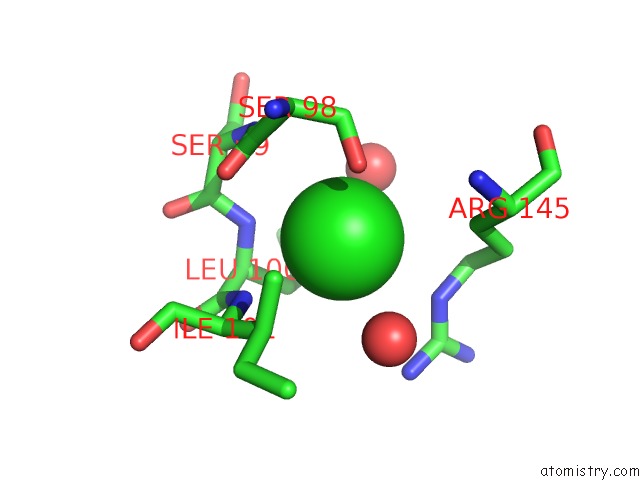
Mono view
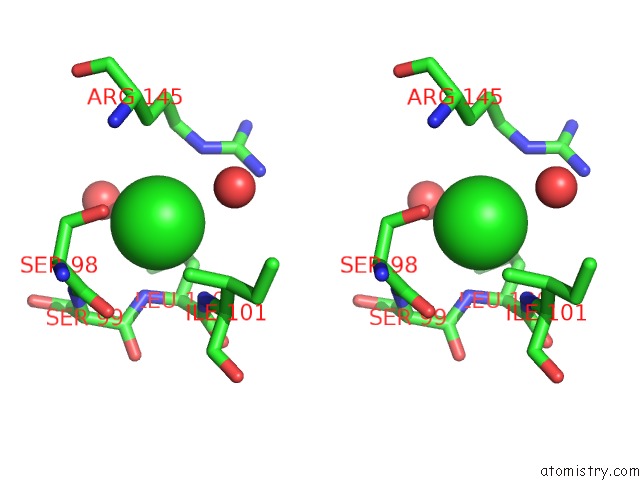
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Crystal Structure of the Effector-Binding Domain of S. Aureus Ccpe within 5.0Å range:
|
Chlorine binding site 2 out of 2 in 4qba
Go back to
Chlorine binding site 2 out
of 2 in the Crystal Structure of the Effector-Binding Domain of S. Aureus Ccpe
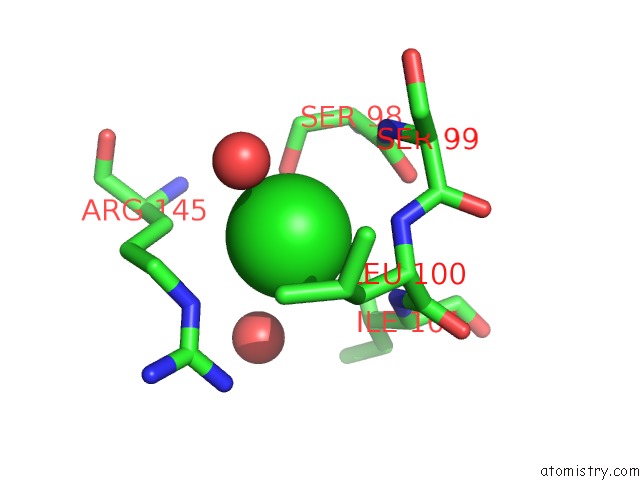
Mono view
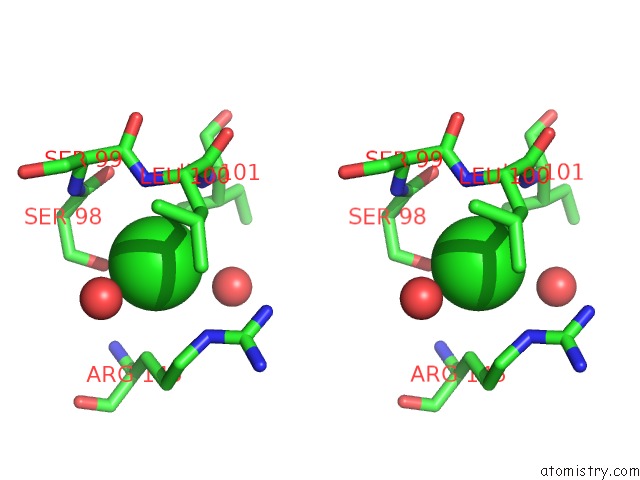
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Crystal Structure of the Effector-Binding Domain of S. Aureus Ccpe within 5.0Å range:
|
Reference:
Y.Ding,
X.Liu,
F.Chen,
H.Di,
B.Xu,
L.Zhou,
X.Deng,
M.Wu,
C.G.Yang,
L.Lan.
Metabolic Sensor Governing Bacterial Virulence in Staphylococcus Aureus. Proc.Natl.Acad.Sci.Usa 2014.
ISSN: ESSN 1091-6490
PubMed: 25368190
DOI: 10.1073/PNAS.1411077111
Page generated: Fri Jul 11 20:44:46 2025
ISSN: ESSN 1091-6490
PubMed: 25368190
DOI: 10.1073/PNAS.1411077111
Last articles
Fe in 7QZHFe in 7QZR
Fe in 7QZG
Fe in 7QZF
Fe in 7QZA
Fe in 7QZE
Fe in 7QSO
Fe in 7QYZ
Fe in 7QYQ
Fe in 7QSN