Chlorine »
PDB 4u3b-4ufx »
4ucc »
Chlorine in PDB 4ucc: N-Terminal Globular Domain of the Rsv Nucleoprotein in Complex with the Nucleoprotein Phosphoprotein Interaction Inhibitor M76
Protein crystallography data
The structure of N-Terminal Globular Domain of the Rsv Nucleoprotein in Complex with the Nucleoprotein Phosphoprotein Interaction Inhibitor M76, PDB code: 4ucc
was solved by
M.Ouizougun-Oubari,
N.Pereira,
B.Tarus,
M.Galloux,
M.-A.Tortorici,
S.Hoos,
B.Baron,
P.England,
F.Bontems,
F.A.Rey,
J.-F.Eleouet,
C.Sizun,
A.Slama-Schwok,
S.Duquerroy,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 15.00 / 2.05 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 34.820, 71.960, 178.170, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 19.14 / 23.58 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the N-Terminal Globular Domain of the Rsv Nucleoprotein in Complex with the Nucleoprotein Phosphoprotein Interaction Inhibitor M76
(pdb code 4ucc). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 4 binding sites of Chlorine where determined in the N-Terminal Globular Domain of the Rsv Nucleoprotein in Complex with the Nucleoprotein Phosphoprotein Interaction Inhibitor M76, PDB code: 4ucc:
Jump to Chlorine binding site number: 1; 2; 3; 4;
In total 4 binding sites of Chlorine where determined in the N-Terminal Globular Domain of the Rsv Nucleoprotein in Complex with the Nucleoprotein Phosphoprotein Interaction Inhibitor M76, PDB code: 4ucc:
Jump to Chlorine binding site number: 1; 2; 3; 4;
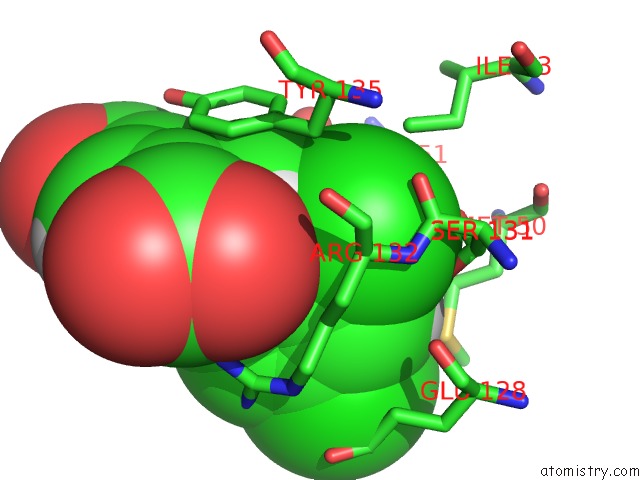
Chlorine binding site 1 out of 4 in 4ucc
Go back to
Chlorine binding site 1 out
of 4 in the N-Terminal Globular Domain of the Rsv Nucleoprotein in Complex with the Nucleoprotein Phosphoprotein Interaction Inhibitor M76
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of N-Terminal Globular Domain of the Rsv Nucleoprotein in Complex with the Nucleoprotein Phosphoprotein Interaction Inhibitor M76 within 5.0Å range:
|
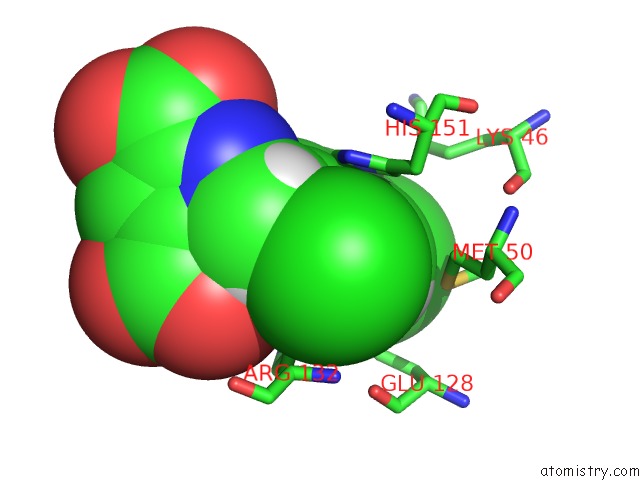
Chlorine binding site 2 out of 4 in 4ucc
Go back to
Chlorine binding site 2 out
of 4 in the N-Terminal Globular Domain of the Rsv Nucleoprotein in Complex with the Nucleoprotein Phosphoprotein Interaction Inhibitor M76
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of N-Terminal Globular Domain of the Rsv Nucleoprotein in Complex with the Nucleoprotein Phosphoprotein Interaction Inhibitor M76 within 5.0Å range:
|
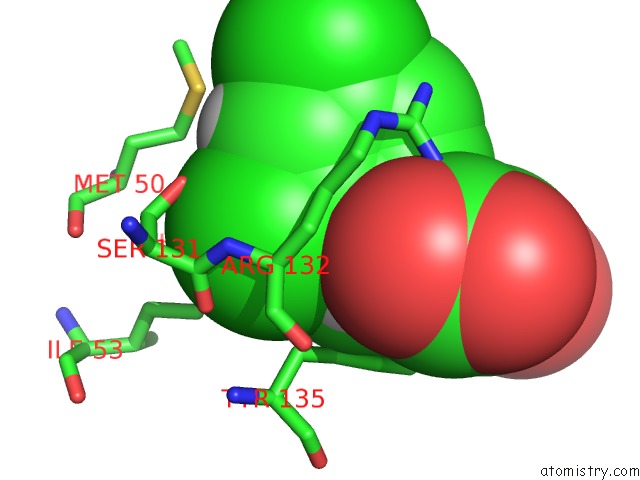
Chlorine binding site 3 out of 4 in 4ucc
Go back to
Chlorine binding site 3 out
of 4 in the N-Terminal Globular Domain of the Rsv Nucleoprotein in Complex with the Nucleoprotein Phosphoprotein Interaction Inhibitor M76
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of N-Terminal Globular Domain of the Rsv Nucleoprotein in Complex with the Nucleoprotein Phosphoprotein Interaction Inhibitor M76 within 5.0Å range:
|
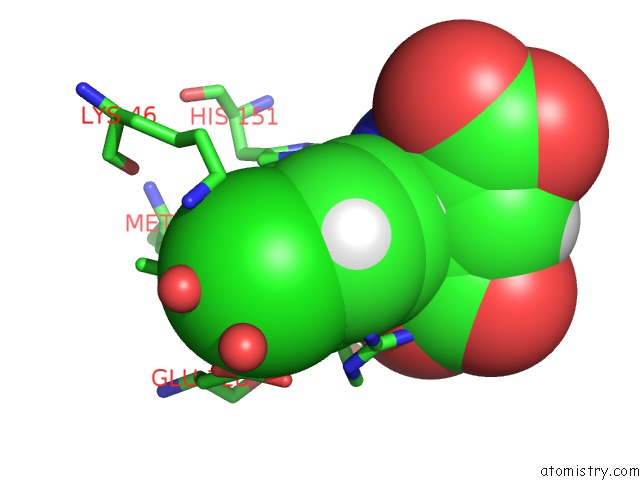
Chlorine binding site 4 out of 4 in 4ucc
Go back to
Chlorine binding site 4 out
of 4 in the N-Terminal Globular Domain of the Rsv Nucleoprotein in Complex with the Nucleoprotein Phosphoprotein Interaction Inhibitor M76
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of N-Terminal Globular Domain of the Rsv Nucleoprotein in Complex with the Nucleoprotein Phosphoprotein Interaction Inhibitor M76 within 5.0Å range:
|
Reference:
M.Ouizougun-Oubari,
N.Pereira,
B.Tarus,
M.Galloux,
S.Lassoued,
J.Fix,
M.A.Tortorici,
S.Hoos,
B.Baron,
P.England,
D.Desmaele,
P.Couvreur,
F.Bontems,
F.A.Rey,
J.F.Eleouet,
C.Sizun,
A.Slama-Schwok,
S.Duquerroy.
A Druggable Pocket at the Nucleocapsid/Phosphoprotein Interaction Site of the Human Respiratory Syncytial Virus. J.Virol. V. 89 11129 2015.
ISSN: ISSN 0022-538X
PubMed: 26246564
DOI: 10.1128/JVI.01612-15
Page generated: Fri Jul 11 22:01:08 2025
ISSN: ISSN 0022-538X
PubMed: 26246564
DOI: 10.1128/JVI.01612-15
Last articles
Mg in 3DHFMg in 3DHD
Mg in 3DIL
Mg in 3DGG
Mg in 3DG7
Mg in 3DGT
Mg in 3DGB
Mg in 3DG6
Mg in 3DG3
Mg in 3DES