Chlorine »
PDB 4uvr-4v3k »
4uwy »
Chlorine in PDB 4uwy: FGFR1 Apo Structure
Enzymatic activity of FGFR1 Apo Structure
All present enzymatic activity of FGFR1 Apo Structure:
2.7.10.1;
2.7.10.1;
Protein crystallography data
The structure of FGFR1 Apo Structure, PDB code: 4uwy
was solved by
N.Thiyagarajan,
T.Bunney,
M.Katan,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 27.815 / 2.30 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 211.837, 49.792, 66.054, 90.00, 107.40, 90.00 |
| R / Rfree (%) | 21.14 / 26.66 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the FGFR1 Apo Structure
(pdb code 4uwy). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 3 binding sites of Chlorine where determined in the FGFR1 Apo Structure, PDB code: 4uwy:
Jump to Chlorine binding site number: 1; 2; 3;
In total 3 binding sites of Chlorine where determined in the FGFR1 Apo Structure, PDB code: 4uwy:
Jump to Chlorine binding site number: 1; 2; 3;
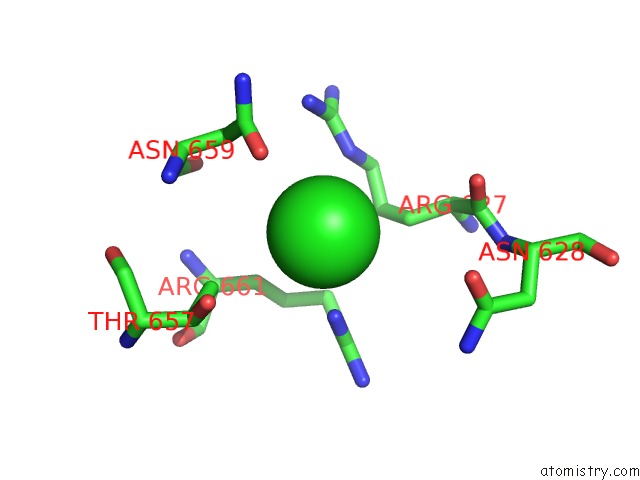
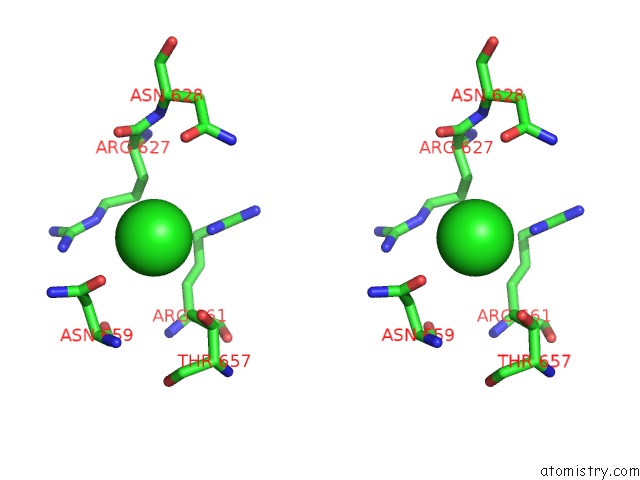
Chlorine binding site 1 out of 3 in 4uwy
Go back to
Chlorine binding site 1 out
of 3 in the FGFR1 Apo Structure
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of FGFR1 Apo Structure within 5.0Å range:
|
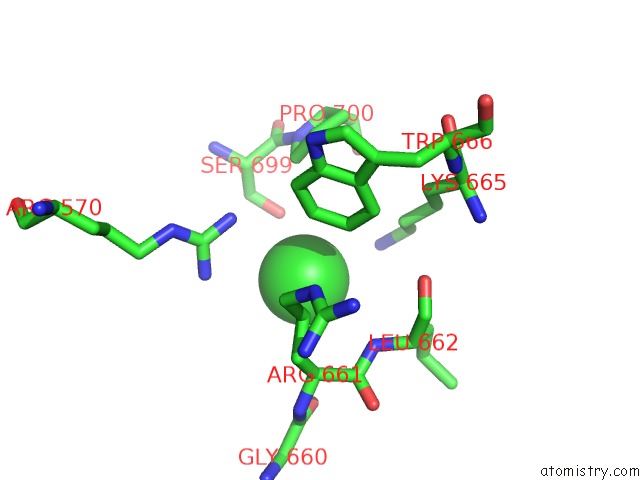
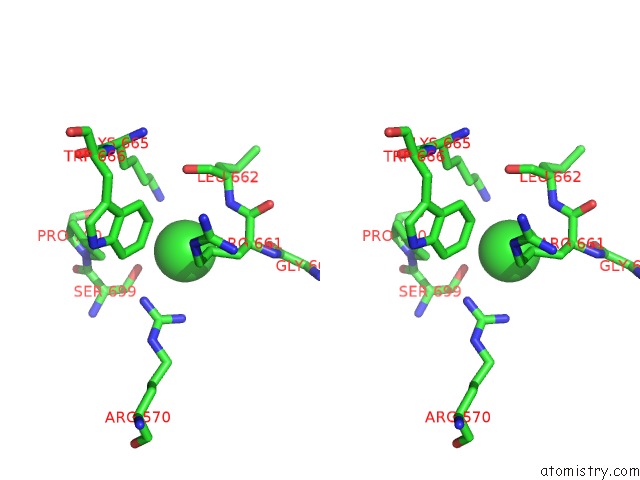
Chlorine binding site 2 out of 3 in 4uwy
Go back to
Chlorine binding site 2 out
of 3 in the FGFR1 Apo Structure
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of FGFR1 Apo Structure within 5.0Å range:
|
Chlorine binding site 3 out of 3 in 4uwy
Go back to
Chlorine binding site 3 out
of 3 in the FGFR1 Apo Structure
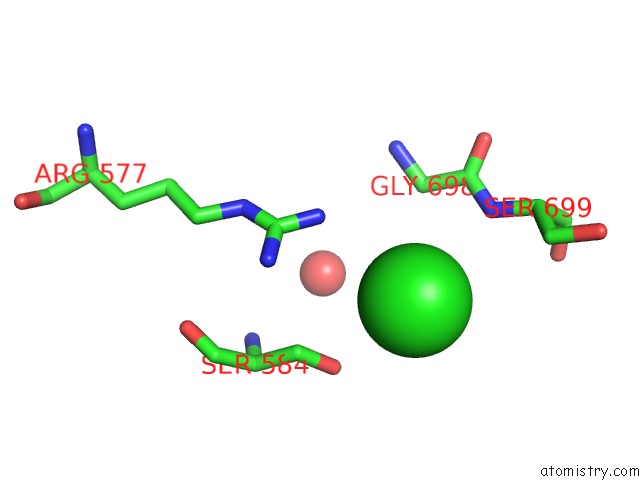
Mono view
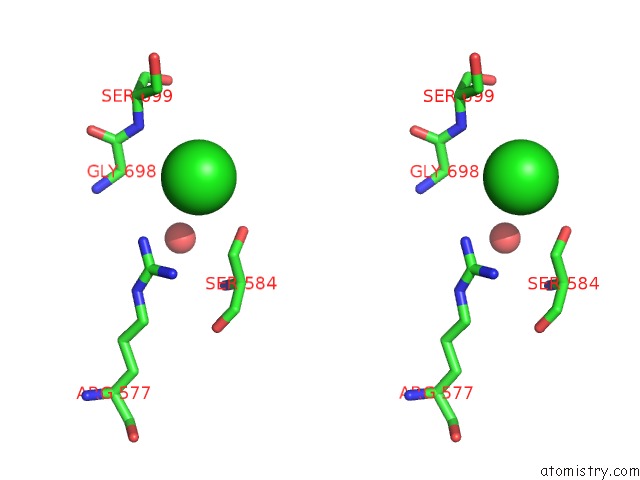
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of FGFR1 Apo Structure within 5.0Å range:
|
Reference:
T.Bunney,
S.Wan,
N.Thiyagarajan,
L.Sutto,
S.V.Williams,
P.Ashford,
H.Koss,
M.A.Knowles,
F.L.Gervasio,
P.V.Coveney,
M.Katan.
The Effect of Mutations on Drug Sensitivity and Kinase Activity of Fibroblast Growth Factor Receptors: A Combined Experimental and Theoretical Study Ebiomedicine 2015.
ISSN: ESSN 2352-3964
DOI: 10.1016/J.EBIOM.2015.02.009
Page generated: Fri Jul 11 22:17:54 2025
ISSN: ESSN 2352-3964
DOI: 10.1016/J.EBIOM.2015.02.009
Last articles
Mg in 5WPMMg in 5WPL
Mg in 5WNS
Mg in 5WNQ
Mg in 5WNR
Mg in 5WNP
Mg in 5WMB
Mg in 5WM8
Mg in 5WNO
Mg in 5WNI