Chlorine »
PDB 5cu8-5d0x »
5cyv »
Chlorine in PDB 5cyv: Crystal Structure of Cour From Rhodococcus Jostii RHA1 Bound to P- Coumaroyl-Coa
Protein crystallography data
The structure of Crystal Structure of Cour From Rhodococcus Jostii RHA1 Bound to P- Coumaroyl-Coa, PDB code: 5cyv
was solved by
P.J.Stogios,
X.Xu,
A.Dong,
A.Savchenko,
A.Joachimiak,
Midwest Center Forstructural Genomics (Mcsg),
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 24.79 / 1.52 |
| Space group | C 2 2 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 64.145, 134.385, 73.336, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 16.1 / 20.2 |
Other elements in 5cyv:
The structure of Crystal Structure of Cour From Rhodococcus Jostii RHA1 Bound to P- Coumaroyl-Coa also contains other interesting chemical elements:
| Magnesium | (Mg) | 3 atoms |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Crystal Structure of Cour From Rhodococcus Jostii RHA1 Bound to P- Coumaroyl-Coa
(pdb code 5cyv). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 3 binding sites of Chlorine where determined in the Crystal Structure of Cour From Rhodococcus Jostii RHA1 Bound to P- Coumaroyl-Coa, PDB code: 5cyv:
Jump to Chlorine binding site number: 1; 2; 3;
In total 3 binding sites of Chlorine where determined in the Crystal Structure of Cour From Rhodococcus Jostii RHA1 Bound to P- Coumaroyl-Coa, PDB code: 5cyv:
Jump to Chlorine binding site number: 1; 2; 3;
Chlorine binding site 1 out of 3 in 5cyv
Go back to
Chlorine binding site 1 out
of 3 in the Crystal Structure of Cour From Rhodococcus Jostii RHA1 Bound to P- Coumaroyl-Coa
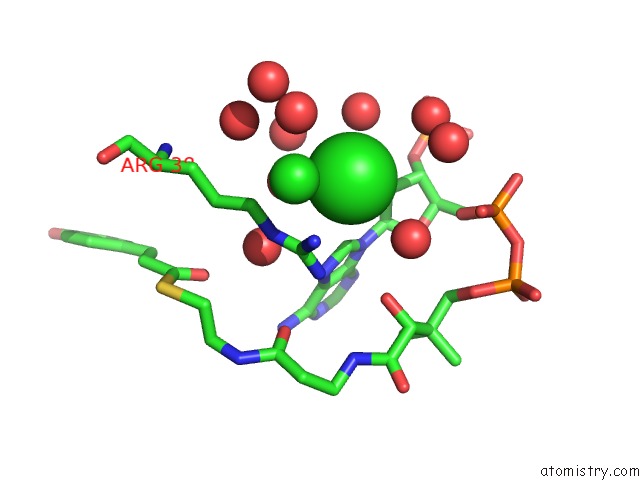
Mono view
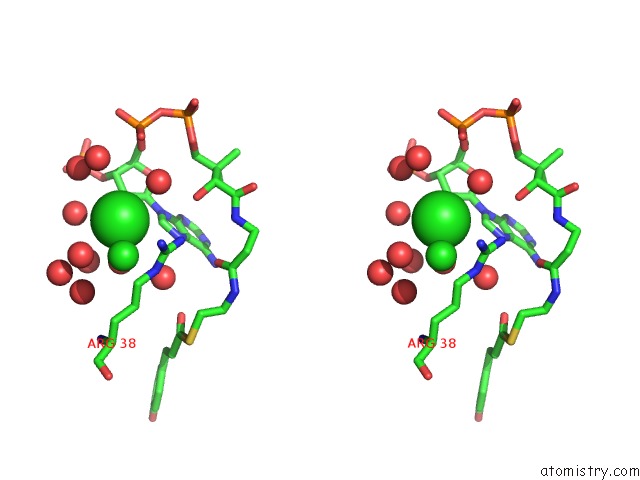
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Crystal Structure of Cour From Rhodococcus Jostii RHA1 Bound to P- Coumaroyl-Coa within 5.0Å range:
|
Chlorine binding site 2 out of 3 in 5cyv
Go back to
Chlorine binding site 2 out
of 3 in the Crystal Structure of Cour From Rhodococcus Jostii RHA1 Bound to P- Coumaroyl-Coa
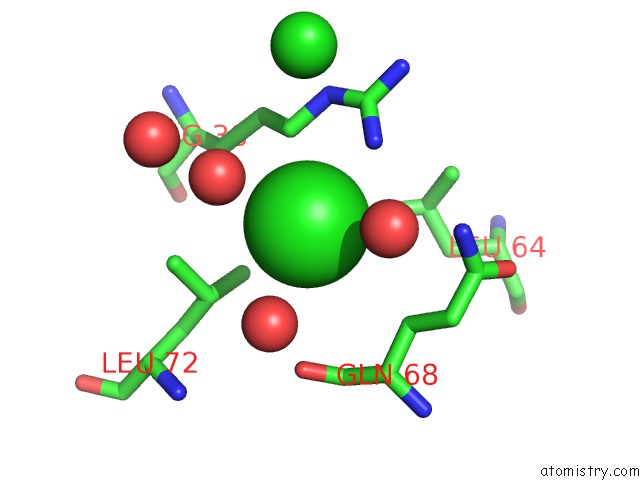
Mono view
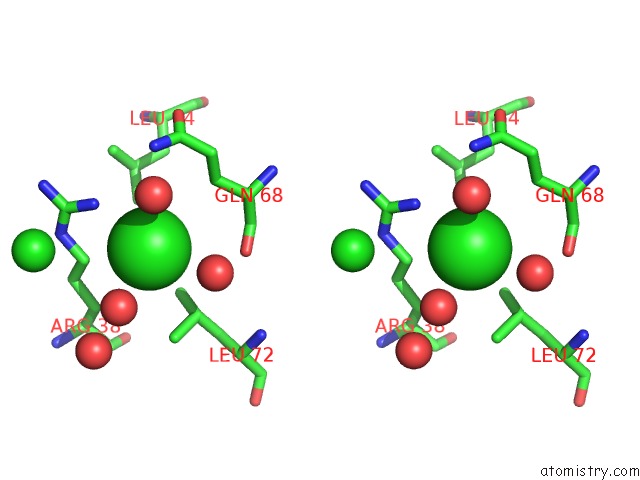
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Crystal Structure of Cour From Rhodococcus Jostii RHA1 Bound to P- Coumaroyl-Coa within 5.0Å range:
|
Chlorine binding site 3 out of 3 in 5cyv
Go back to
Chlorine binding site 3 out
of 3 in the Crystal Structure of Cour From Rhodococcus Jostii RHA1 Bound to P- Coumaroyl-Coa
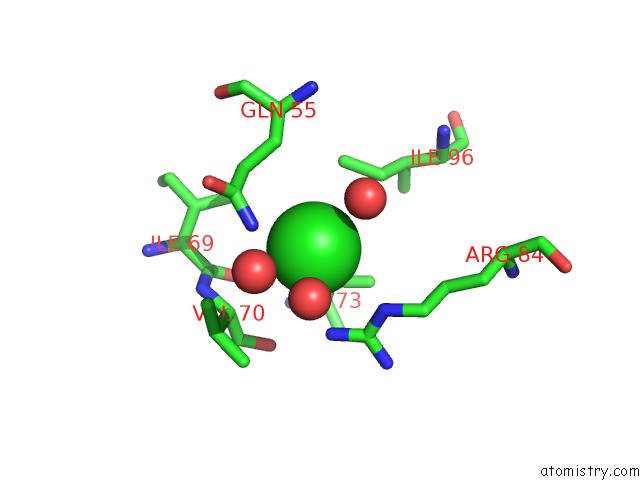
Mono view
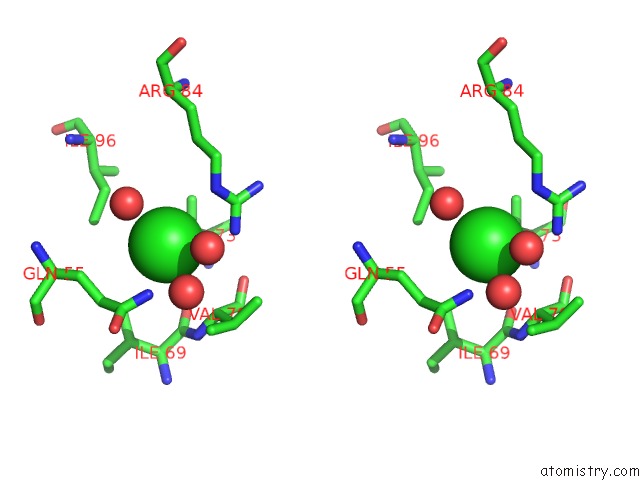
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Crystal Structure of Cour From Rhodococcus Jostii RHA1 Bound to P- Coumaroyl-Coa within 5.0Å range:
|
Reference:
H.Otani,
P.J.Stogios,
X.Xu,
B.Nocek,
S.N.Li,
A.Savchenko,
L.D.Eltis.
The Activity of Cour, A Marr Family Transcriptional Regulator, Is Modulated Through A Novel Molecular Mechanism. Nucleic Acids Res. V. 44 595 2016.
ISSN: ESSN 1362-4962
PubMed: 26400178
DOI: 10.1093/NAR/GKV955
Page generated: Sat Jul 12 01:00:14 2025
ISSN: ESSN 1362-4962
PubMed: 26400178
DOI: 10.1093/NAR/GKV955
Last articles
K in 9EIEK in 9EAF
K in 9DWN
K in 9E4V
K in 9DKF
K in 9DTR
K in 9DXX
K in 9DKL
K in 9DQP
K in 9DSB