Chlorine »
PDB 5d0z-5da3 »
5d2h »
Chlorine in PDB 5d2h: 4-Oxalocrotonate Decarboxylase From Pseudomonas Putida G7 - Complexed with Magnesium and Alpha-Ketoglutarate
Enzymatic activity of 4-Oxalocrotonate Decarboxylase From Pseudomonas Putida G7 - Complexed with Magnesium and Alpha-Ketoglutarate
All present enzymatic activity of 4-Oxalocrotonate Decarboxylase From Pseudomonas Putida G7 - Complexed with Magnesium and Alpha-Ketoglutarate:
4.1.1.77;
4.1.1.77;
Protein crystallography data
The structure of 4-Oxalocrotonate Decarboxylase From Pseudomonas Putida G7 - Complexed with Magnesium and Alpha-Ketoglutarate, PDB code: 5d2h
was solved by
S.L.Guimaraes,
R.A.P.Nagem,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 42.27 / 1.94 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 43.741, 44.904, 125.318, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 16.3 / 23.1 |
Other elements in 5d2h:
The structure of 4-Oxalocrotonate Decarboxylase From Pseudomonas Putida G7 - Complexed with Magnesium and Alpha-Ketoglutarate also contains other interesting chemical elements:
| Magnesium | (Mg) | 1 atom |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the 4-Oxalocrotonate Decarboxylase From Pseudomonas Putida G7 - Complexed with Magnesium and Alpha-Ketoglutarate
(pdb code 5d2h). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total only one binding site of Chlorine was determined in the 4-Oxalocrotonate Decarboxylase From Pseudomonas Putida G7 - Complexed with Magnesium and Alpha-Ketoglutarate, PDB code: 5d2h:
In total only one binding site of Chlorine was determined in the 4-Oxalocrotonate Decarboxylase From Pseudomonas Putida G7 - Complexed with Magnesium and Alpha-Ketoglutarate, PDB code: 5d2h:
Chlorine binding site 1 out of 1 in 5d2h
Go back to
Chlorine binding site 1 out
of 1 in the 4-Oxalocrotonate Decarboxylase From Pseudomonas Putida G7 - Complexed with Magnesium and Alpha-Ketoglutarate
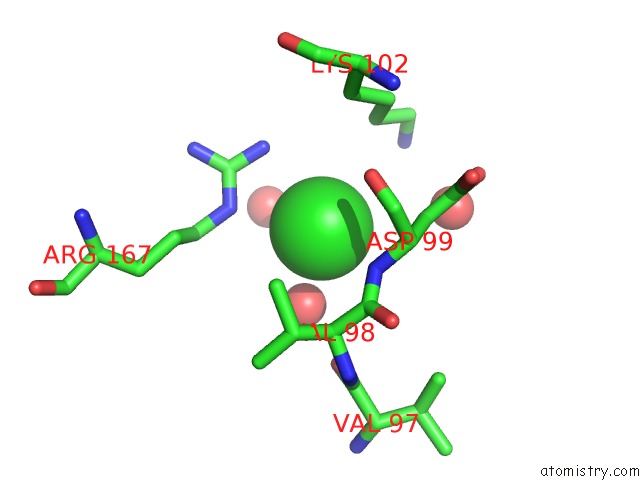
Mono view
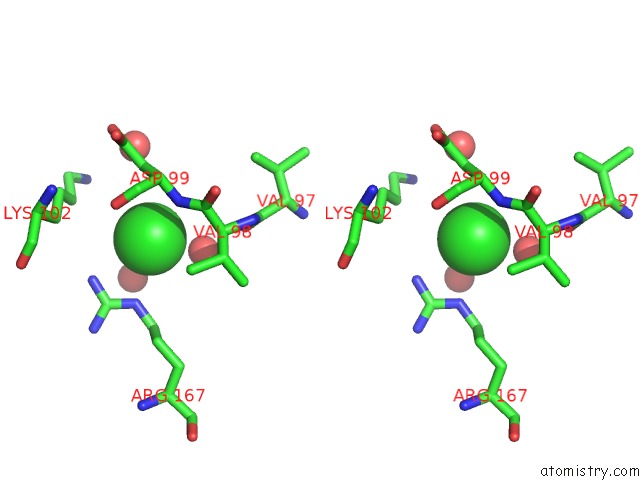
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of 4-Oxalocrotonate Decarboxylase From Pseudomonas Putida G7 - Complexed with Magnesium and Alpha-Ketoglutarate within 5.0Å range:
|
Reference:
S.L.Guimaraes,
J.B.Coitinho,
D.M.Costa,
S.S.Araujo,
C.P.Whitman,
R.A.Nagem.
Crystal Structures of Apo and Liganded 4-Oxalocrotonate Decarboxylase Uncover A Structural Basis For the Metal-Assisted Decarboxylation of A Vinylogous Beta-Keto Acid. Biochemistry V. 55 2632 2016.
ISSN: ISSN 0006-2960
PubMed: 27082660
DOI: 10.1021/ACS.BIOCHEM.6B00050
Page generated: Sat Jul 12 01:05:56 2025
ISSN: ISSN 0006-2960
PubMed: 27082660
DOI: 10.1021/ACS.BIOCHEM.6B00050
Last articles
K in 9DITK in 9DJV
K in 9DIG
K in 9DID
K in 9DIC
K in 9DI8
K in 9DIB
K in 9DE5
K in 9D5W
K in 9D19