Chlorine »
PDB 5e20-5eae »
5e7c »
Chlorine in PDB 5e7c: Macromolecular Diffractive Imaging Using Imperfect Crystals - Bragg Data
Enzymatic activity of Macromolecular Diffractive Imaging Using Imperfect Crystals - Bragg Data
All present enzymatic activity of Macromolecular Diffractive Imaging Using Imperfect Crystals - Bragg Data:
1.10.3.9;
1.10.3.9;
Protein crystallography data
The structure of Macromolecular Diffractive Imaging Using Imperfect Crystals - Bragg Data, PDB code: 5e7c
was solved by
K.Ayyer,
O.Yefanov,
D.Oberthuer,
S.Roy-Chowdhury,
L.Galli,
V.Mariani,
S.Basu,
J.Coe,
C.E.Conrad,
R.Fromme,
A.Schaffner,
K.Doerner,
D.James,
C.Kupitz,
M.Metz,
G.Nelson,
P.L.Xavier,
K.R.Beyerlein,
M.Schmidt,
I.Sarrou,
J.C.H.Spence,
U.Weierstall,
T.A.White,
J.-H.Yang,
Y.Zhao,
M.Liang,
A.Aquila,
M.S.Hunter,
J.S.Robinson,
J.E.Koglin,
S.Boutet,
P.Fromme,
A.Barty,
H.N.Chapman,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 29.93 / 4.50 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 133.250, 226.260, 307.090, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 24.7 / 27.5 |
Other elements in 5e7c:
The structure of Macromolecular Diffractive Imaging Using Imperfect Crystals - Bragg Data also contains other interesting chemical elements:
| Magnesium | (Mg) | 74 atoms |
| Manganese | (Mn) | 8 atoms |
| Iron | (Fe) | 6 atoms |
| Calcium | (Ca) | 8 atoms |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Macromolecular Diffractive Imaging Using Imperfect Crystals - Bragg Data
(pdb code 5e7c). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 6 binding sites of Chlorine where determined in the Macromolecular Diffractive Imaging Using Imperfect Crystals - Bragg Data, PDB code: 5e7c:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5; 6;
In total 6 binding sites of Chlorine where determined in the Macromolecular Diffractive Imaging Using Imperfect Crystals - Bragg Data, PDB code: 5e7c:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5; 6;
Chlorine binding site 1 out of 6 in 5e7c
Go back to
Chlorine binding site 1 out
of 6 in the Macromolecular Diffractive Imaging Using Imperfect Crystals - Bragg Data
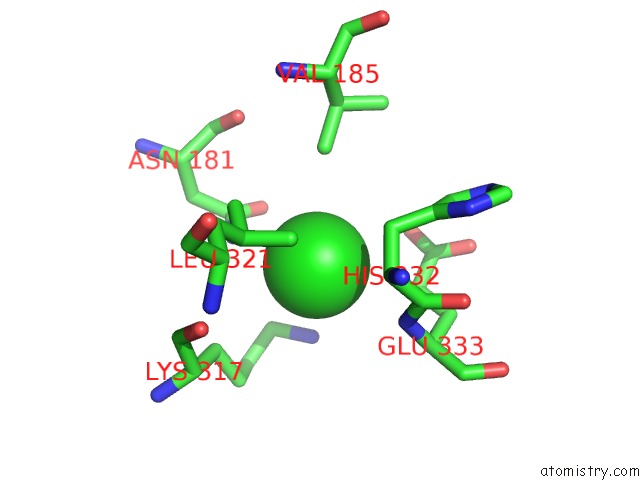
Mono view
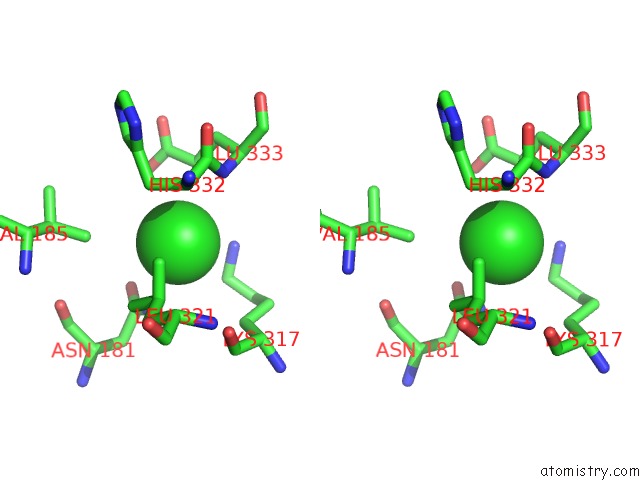
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Macromolecular Diffractive Imaging Using Imperfect Crystals - Bragg Data within 5.0Å range:
|
Chlorine binding site 2 out of 6 in 5e7c
Go back to
Chlorine binding site 2 out
of 6 in the Macromolecular Diffractive Imaging Using Imperfect Crystals - Bragg Data
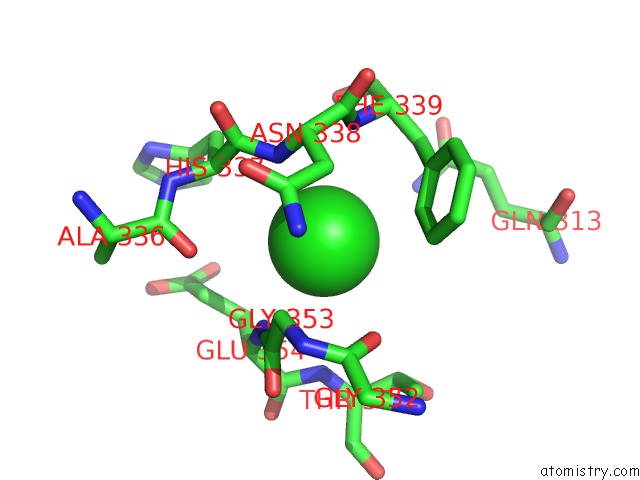
Mono view
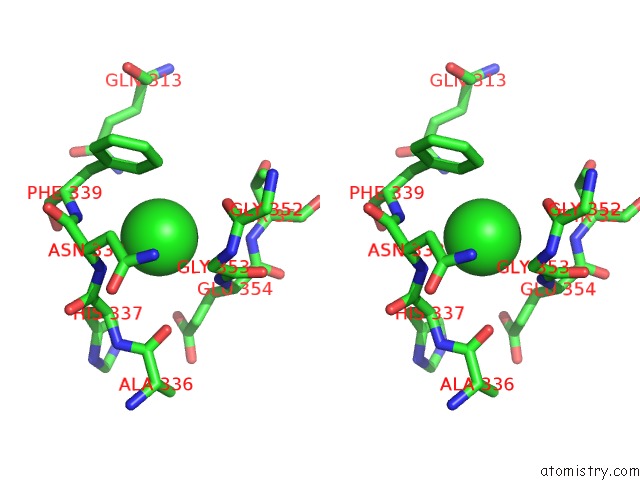
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Macromolecular Diffractive Imaging Using Imperfect Crystals - Bragg Data within 5.0Å range:
|
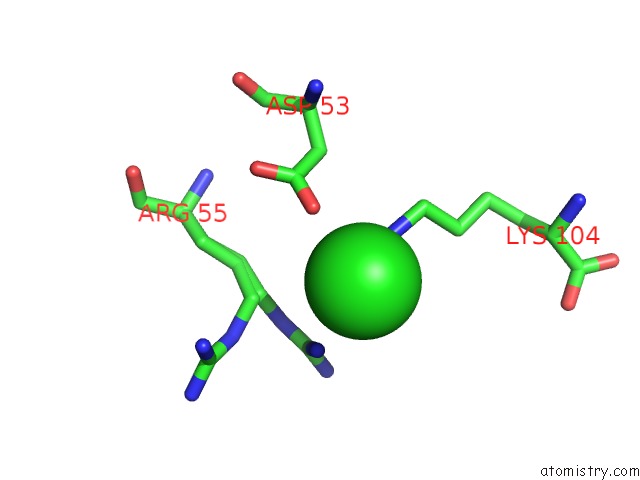
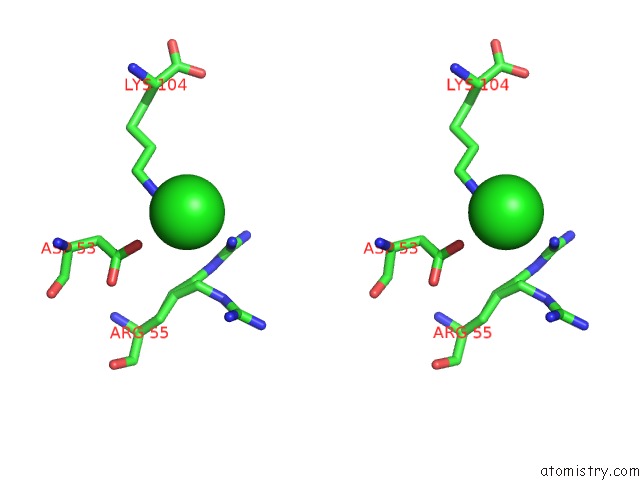
Chlorine binding site 3 out of 6 in 5e7c
Go back to
Chlorine binding site 3 out
of 6 in the Macromolecular Diffractive Imaging Using Imperfect Crystals - Bragg Data
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Macromolecular Diffractive Imaging Using Imperfect Crystals - Bragg Data within 5.0Å range:
|
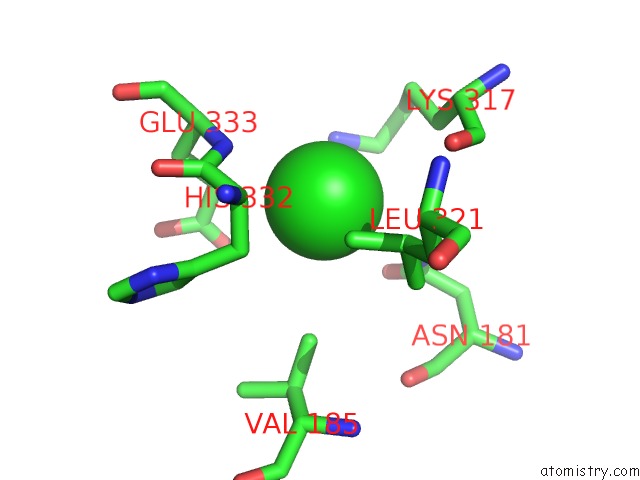
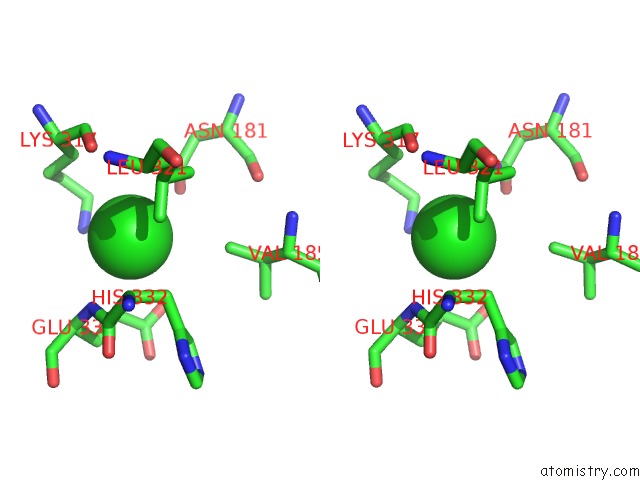
Chlorine binding site 4 out of 6 in 5e7c
Go back to
Chlorine binding site 4 out
of 6 in the Macromolecular Diffractive Imaging Using Imperfect Crystals - Bragg Data
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of Macromolecular Diffractive Imaging Using Imperfect Crystals - Bragg Data within 5.0Å range:
|
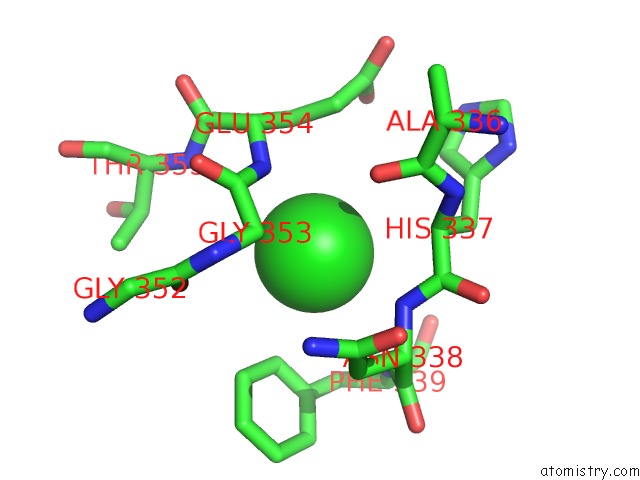
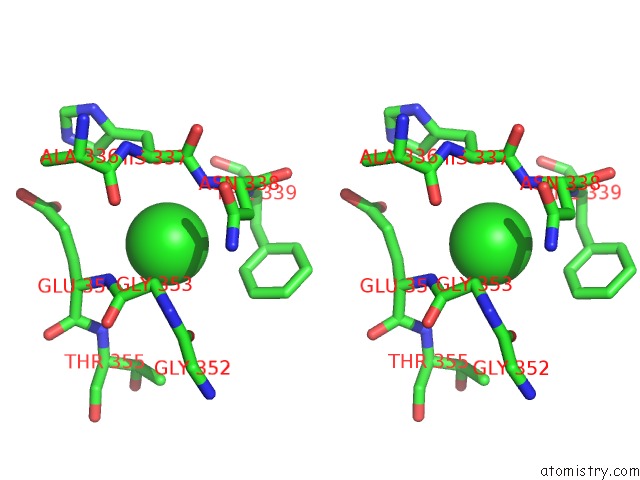
Chlorine binding site 5 out of 6 in 5e7c
Go back to
Chlorine binding site 5 out
of 6 in the Macromolecular Diffractive Imaging Using Imperfect Crystals - Bragg Data
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 5 of Macromolecular Diffractive Imaging Using Imperfect Crystals - Bragg Data within 5.0Å range:
|
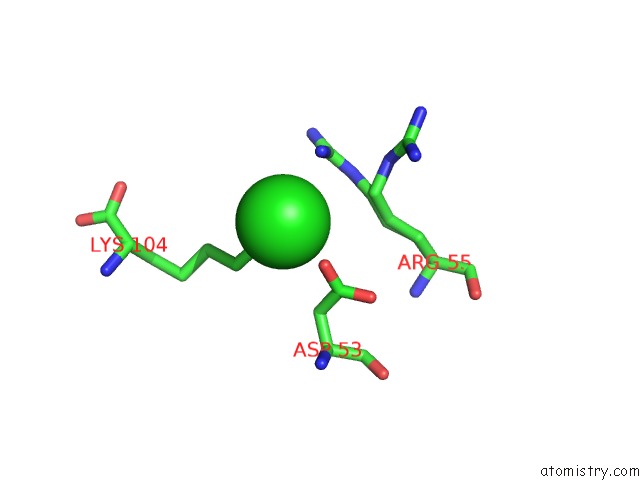
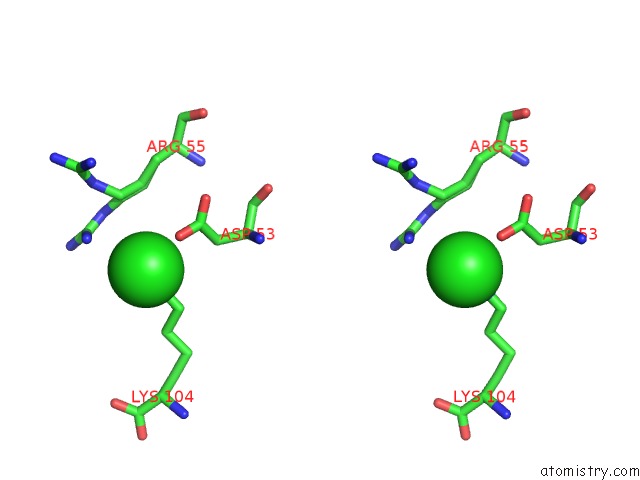
Chlorine binding site 6 out of 6 in 5e7c
Go back to
Chlorine binding site 6 out
of 6 in the Macromolecular Diffractive Imaging Using Imperfect Crystals - Bragg Data
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 6 of Macromolecular Diffractive Imaging Using Imperfect Crystals - Bragg Data within 5.0Å range:
|
Reference:
K.Ayyer,
O.M.Yefanov,
D.Oberthur,
S.Roy-Chowdhury,
L.Galli,
V.Mariani,
S.Basu,
J.Coe,
C.E.Conrad,
R.Fromme,
A.Schaffer,
K.Dorner,
D.James,
C.Kupitz,
M.Metz,
G.Nelson,
P.L.Xavier,
K.R.Beyerlein,
M.Schmidt,
I.Sarrou,
J.C.Spence,
U.Weierstall,
T.A.White,
J.H.Yang,
Y.Zhao,
M.Liang,
A.Aquila,
M.S.Hunter,
J.S.Robinson,
J.E.Koglin,
S.Boutet,
P.Fromme,
A.Barty,
H.N.Chapman.
Macromolecular Diffractive Imaging Using Imperfect Crystals. Nature V. 530 202 2016.
ISSN: ESSN 1476-4687
PubMed: 26863980
DOI: 10.1038/NATURE16949
Page generated: Sat Jul 12 01:32:54 2025
ISSN: ESSN 1476-4687
PubMed: 26863980
DOI: 10.1038/NATURE16949
Last articles
I in 4OHMI in 4OHK
I in 4OEK
I in 4OEW
I in 4OC5
I in 4OC4
I in 4OC3
I in 4OC1
I in 4OC2
I in 4NHB