Chlorine »
PDB 5f03-5f6q »
5f47 »
Chlorine in PDB 5f47: Crystal Structure of An Aminoglycoside Acetyltransferase Meta-AAC0020 From An Uncultured Soil Metagenomic Sample in Complex with Trehalose
Protein crystallography data
The structure of Crystal Structure of An Aminoglycoside Acetyltransferase Meta-AAC0020 From An Uncultured Soil Metagenomic Sample in Complex with Trehalose, PDB code: 5f47
was solved by
Z.Xu,
T.Skarina,
Z.Wawrzak,
P.J.Stogios,
V.Yim,
A.Savchenko,
W.F.Anderson,
Center For Structural Genomics Of Infectious Diseases (Csgid),
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 26.98 / 1.50 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 140.001, 53.954, 46.023, 90.00, 105.19, 90.00 |
| R / Rfree (%) | 15.6 / 17.8 |
Other elements in 5f47:
The structure of Crystal Structure of An Aminoglycoside Acetyltransferase Meta-AAC0020 From An Uncultured Soil Metagenomic Sample in Complex with Trehalose also contains other interesting chemical elements:
| Calcium | (Ca) | 8 atoms |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Crystal Structure of An Aminoglycoside Acetyltransferase Meta-AAC0020 From An Uncultured Soil Metagenomic Sample in Complex with Trehalose
(pdb code 5f47). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 2 binding sites of Chlorine where determined in the Crystal Structure of An Aminoglycoside Acetyltransferase Meta-AAC0020 From An Uncultured Soil Metagenomic Sample in Complex with Trehalose, PDB code: 5f47:
Jump to Chlorine binding site number: 1; 2;
In total 2 binding sites of Chlorine where determined in the Crystal Structure of An Aminoglycoside Acetyltransferase Meta-AAC0020 From An Uncultured Soil Metagenomic Sample in Complex with Trehalose, PDB code: 5f47:
Jump to Chlorine binding site number: 1; 2;
Chlorine binding site 1 out of 2 in 5f47
Go back to
Chlorine binding site 1 out
of 2 in the Crystal Structure of An Aminoglycoside Acetyltransferase Meta-AAC0020 From An Uncultured Soil Metagenomic Sample in Complex with Trehalose
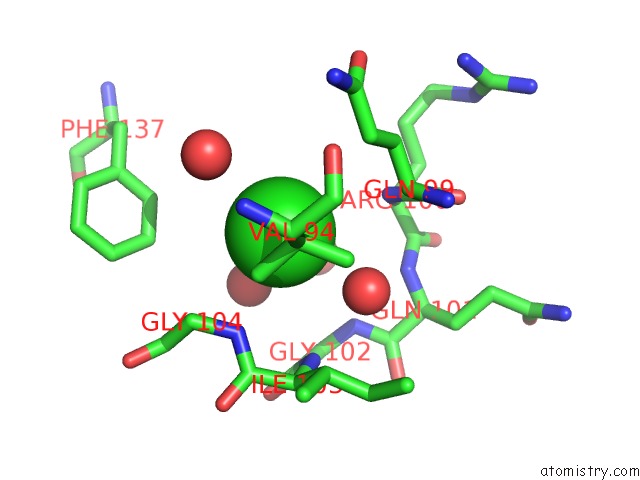
Mono view
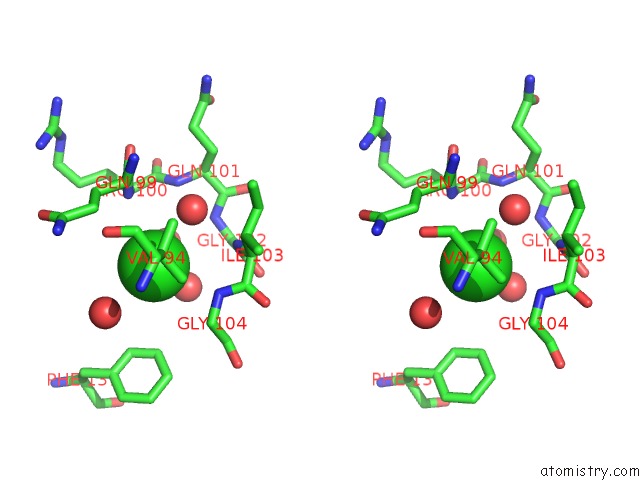
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Crystal Structure of An Aminoglycoside Acetyltransferase Meta-AAC0020 From An Uncultured Soil Metagenomic Sample in Complex with Trehalose within 5.0Å range:
|
Chlorine binding site 2 out of 2 in 5f47
Go back to
Chlorine binding site 2 out
of 2 in the Crystal Structure of An Aminoglycoside Acetyltransferase Meta-AAC0020 From An Uncultured Soil Metagenomic Sample in Complex with Trehalose
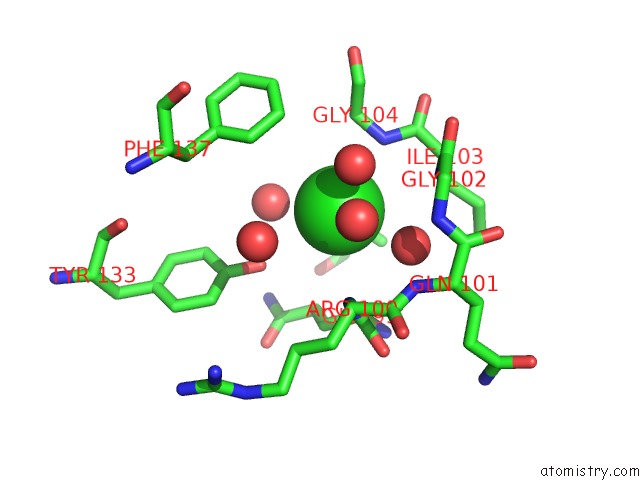
Mono view
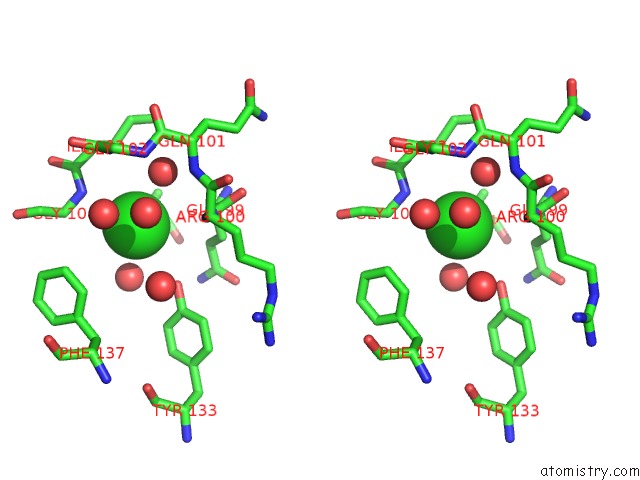
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Crystal Structure of An Aminoglycoside Acetyltransferase Meta-AAC0020 From An Uncultured Soil Metagenomic Sample in Complex with Trehalose within 5.0Å range:
|
Reference:
Z.Xu,
P.J.Stogios,
A.T.Quaile,
K.J.Forsberg,
S.Patel,
T.Skarina,
S.Houliston,
C.Arrowsmith,
G.Dantas,
A.Savchenko.
Structural and Functional Survey of Environmental Aminoglycoside Acetyltransferases Reveals Functionality of Resistance Enzymes. Acs Infect Dis V. 3 653 2017.
ISSN: ESSN 2373-8227
PubMed: 28756664
DOI: 10.1021/ACSINFECDIS.7B00068
Page generated: Sat Jul 12 01:54:39 2025
ISSN: ESSN 2373-8227
PubMed: 28756664
DOI: 10.1021/ACSINFECDIS.7B00068
Last articles
F in 8SHDF in 8SHA
F in 8SGL
F in 8SH9
F in 8SHC
F in 8SGC
F in 8SG9
F in 8SGI
F in 8SG8
F in 8SFF