Chlorine »
PDB 5nd0-5nkd »
5ngj »
Chlorine in PDB 5ngj: Crystal Structure of PB6, Major Tail Tube Protein of Bacteriophage T5
Protein crystallography data
The structure of Crystal Structure of PB6, Major Tail Tube Protein of Bacteriophage T5, PDB code: 5ngj
was solved by
C.-A.Arnaud,
G.Effantin,
C.Vives,
S.Engilberge,
M.Bacia,
P.Boulanger,
E.Girard,
G.Schoehn,
C.Breyton,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 44.00 / 2.20 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 76.790, 115.081, 83.366, 90.00, 111.88, 90.00 |
| R / Rfree (%) | 20.5 / 23.5 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Crystal Structure of PB6, Major Tail Tube Protein of Bacteriophage T5
(pdb code 5ngj). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 5 binding sites of Chlorine where determined in the Crystal Structure of PB6, Major Tail Tube Protein of Bacteriophage T5, PDB code: 5ngj:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5;
In total 5 binding sites of Chlorine where determined in the Crystal Structure of PB6, Major Tail Tube Protein of Bacteriophage T5, PDB code: 5ngj:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5;
Chlorine binding site 1 out of 5 in 5ngj
Go back to
Chlorine binding site 1 out
of 5 in the Crystal Structure of PB6, Major Tail Tube Protein of Bacteriophage T5
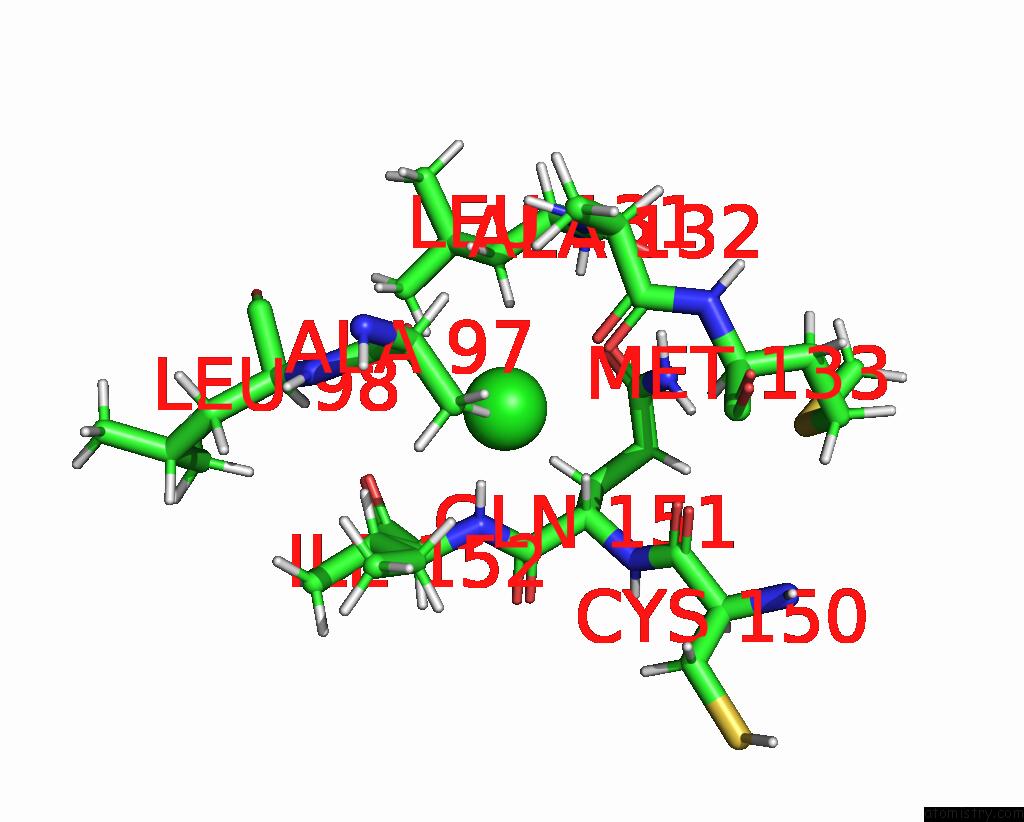
Mono view
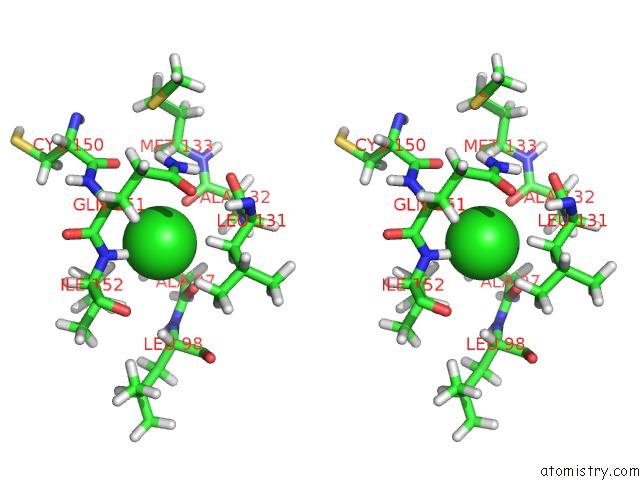
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Crystal Structure of PB6, Major Tail Tube Protein of Bacteriophage T5 within 5.0Å range:
|
Chlorine binding site 2 out of 5 in 5ngj
Go back to
Chlorine binding site 2 out
of 5 in the Crystal Structure of PB6, Major Tail Tube Protein of Bacteriophage T5
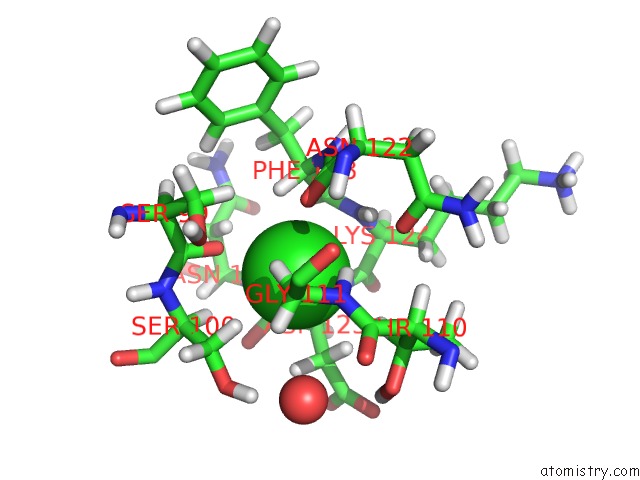
Mono view
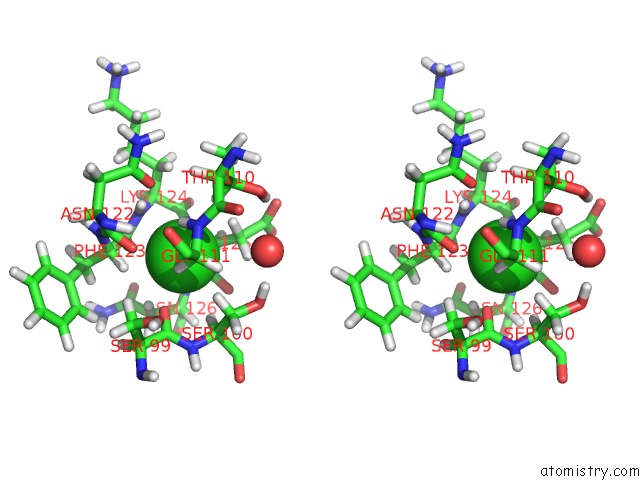
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Crystal Structure of PB6, Major Tail Tube Protein of Bacteriophage T5 within 5.0Å range:
|
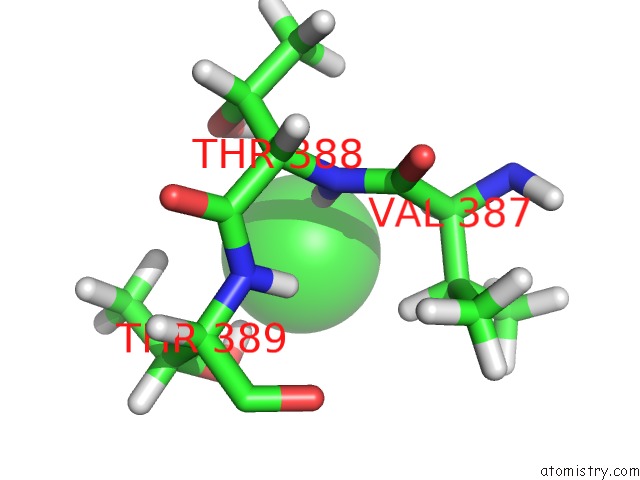
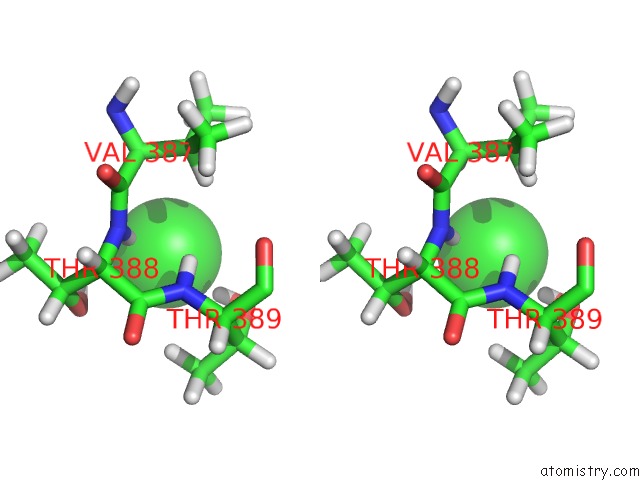
Chlorine binding site 3 out of 5 in 5ngj
Go back to
Chlorine binding site 3 out
of 5 in the Crystal Structure of PB6, Major Tail Tube Protein of Bacteriophage T5
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Crystal Structure of PB6, Major Tail Tube Protein of Bacteriophage T5 within 5.0Å range:
|
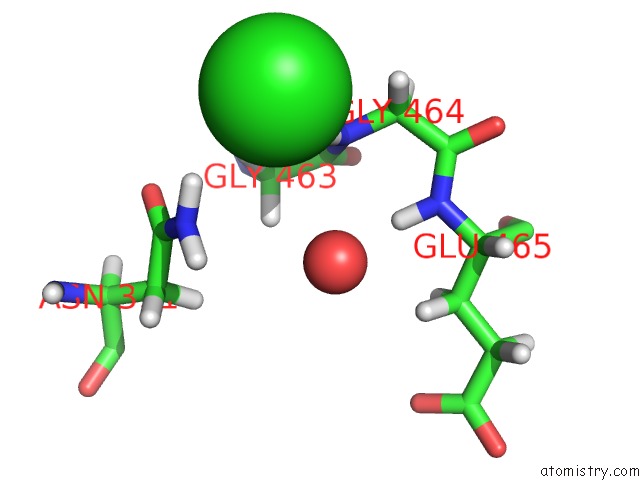
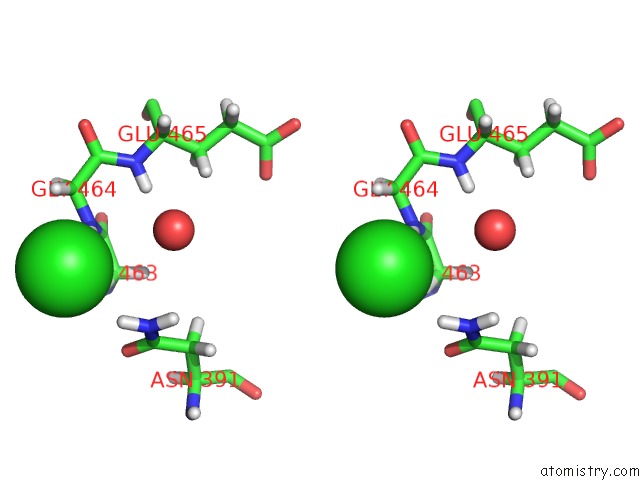
Chlorine binding site 4 out of 5 in 5ngj
Go back to
Chlorine binding site 4 out
of 5 in the Crystal Structure of PB6, Major Tail Tube Protein of Bacteriophage T5
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of Crystal Structure of PB6, Major Tail Tube Protein of Bacteriophage T5 within 5.0Å range:
|
Chlorine binding site 5 out of 5 in 5ngj
Go back to
Chlorine binding site 5 out
of 5 in the Crystal Structure of PB6, Major Tail Tube Protein of Bacteriophage T5
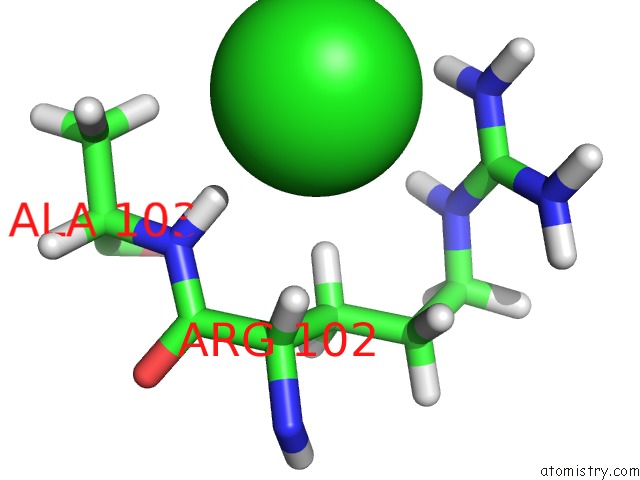
Mono view
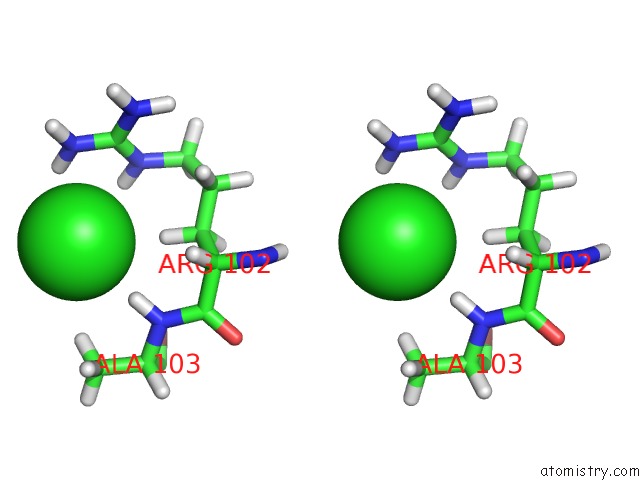
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 5 of Crystal Structure of PB6, Major Tail Tube Protein of Bacteriophage T5 within 5.0Å range:
|
Reference:
C.A.Arnaud,
G.Effantin,
C.Vives,
S.Engilberge,
M.Bacia,
P.Boulanger,
E.Girard,
G.Schoehn,
C.Breyton.
Bacteriophage T5 Tail Tube Structure Suggests A Trigger Mechanism For Siphoviridae Dna Ejection. Nat Commun V. 8 1953 2017.
ISSN: ESSN 2041-1723
PubMed: 29209037
DOI: 10.1038/S41467-017-02049-3
Page generated: Sat Jul 12 06:06:22 2025
ISSN: ESSN 2041-1723
PubMed: 29209037
DOI: 10.1038/S41467-017-02049-3
Last articles
Mg in 7BRSMg in 7BPI
Mg in 7BOG
Mg in 7BPH
Mg in 7BOF
Mg in 7BOE
Mg in 7BGI
Mg in 7BLX
Mg in 7BLZ
Mg in 7BOD