Chlorine »
PDB 5upg-5uwx »
5upz »
Chlorine in PDB 5upz: Hiv-1 Wild Type Protease with Grl-0518A , An Isophthalamide-Derived P2-P3 Ligand with the Para-Hydoxymethyl Sulfonamide Isostere As the P2' Group
Protein crystallography data
The structure of Hiv-1 Wild Type Protease with Grl-0518A , An Isophthalamide-Derived P2-P3 Ligand with the Para-Hydoxymethyl Sulfonamide Isostere As the P2' Group, PDB code: 5upz
was solved by
Y.-F.Wang,
J.Agniswamy,
I.T.Weber,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 50.00 / 1.27 |
| Space group | P 21 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 58.175, 86.418, 45.828, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 14.5 / 19.8 |
Other elements in 5upz:
The structure of Hiv-1 Wild Type Protease with Grl-0518A , An Isophthalamide-Derived P2-P3 Ligand with the Para-Hydoxymethyl Sulfonamide Isostere As the P2' Group also contains other interesting chemical elements:
| Sodium | (Na) | 2 atoms |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Hiv-1 Wild Type Protease with Grl-0518A , An Isophthalamide-Derived P2-P3 Ligand with the Para-Hydoxymethyl Sulfonamide Isostere As the P2' Group
(pdb code 5upz). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 3 binding sites of Chlorine where determined in the Hiv-1 Wild Type Protease with Grl-0518A , An Isophthalamide-Derived P2-P3 Ligand with the Para-Hydoxymethyl Sulfonamide Isostere As the P2' Group, PDB code: 5upz:
Jump to Chlorine binding site number: 1; 2; 3;
In total 3 binding sites of Chlorine where determined in the Hiv-1 Wild Type Protease with Grl-0518A , An Isophthalamide-Derived P2-P3 Ligand with the Para-Hydoxymethyl Sulfonamide Isostere As the P2' Group, PDB code: 5upz:
Jump to Chlorine binding site number: 1; 2; 3;
Chlorine binding site 1 out of 3 in 5upz
Go back to
Chlorine binding site 1 out
of 3 in the Hiv-1 Wild Type Protease with Grl-0518A , An Isophthalamide-Derived P2-P3 Ligand with the Para-Hydoxymethyl Sulfonamide Isostere As the P2' Group
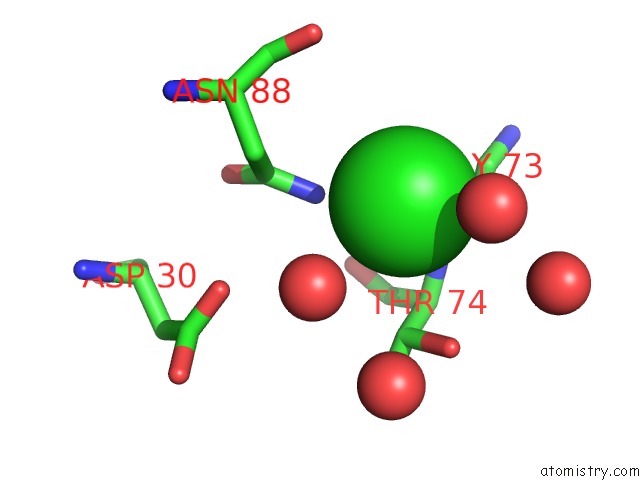
Mono view
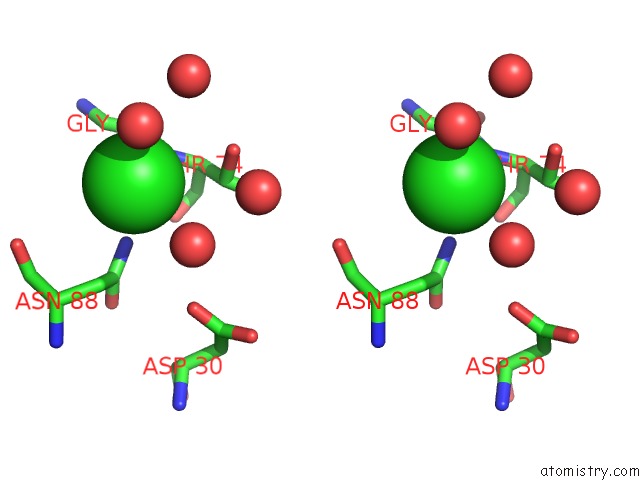
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Hiv-1 Wild Type Protease with Grl-0518A , An Isophthalamide-Derived P2-P3 Ligand with the Para-Hydoxymethyl Sulfonamide Isostere As the P2' Group within 5.0Å range:
|
Chlorine binding site 2 out of 3 in 5upz
Go back to
Chlorine binding site 2 out
of 3 in the Hiv-1 Wild Type Protease with Grl-0518A , An Isophthalamide-Derived P2-P3 Ligand with the Para-Hydoxymethyl Sulfonamide Isostere As the P2' Group
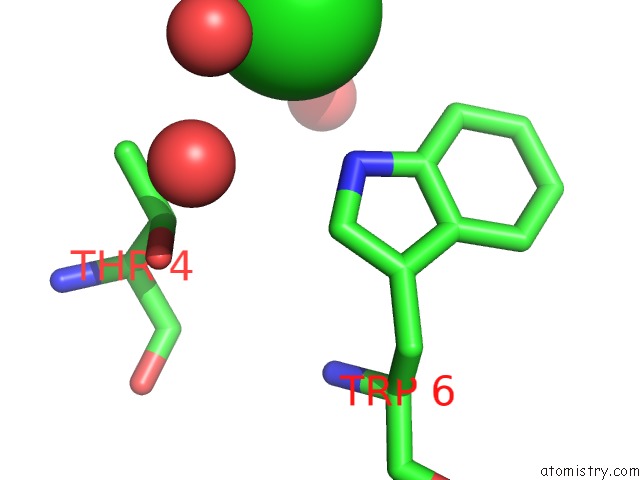
Mono view
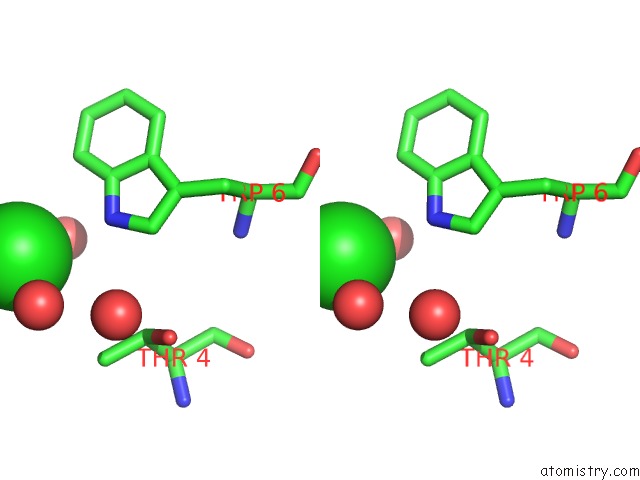
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Hiv-1 Wild Type Protease with Grl-0518A , An Isophthalamide-Derived P2-P3 Ligand with the Para-Hydoxymethyl Sulfonamide Isostere As the P2' Group within 5.0Å range:
|
Chlorine binding site 3 out of 3 in 5upz
Go back to
Chlorine binding site 3 out
of 3 in the Hiv-1 Wild Type Protease with Grl-0518A , An Isophthalamide-Derived P2-P3 Ligand with the Para-Hydoxymethyl Sulfonamide Isostere As the P2' Group
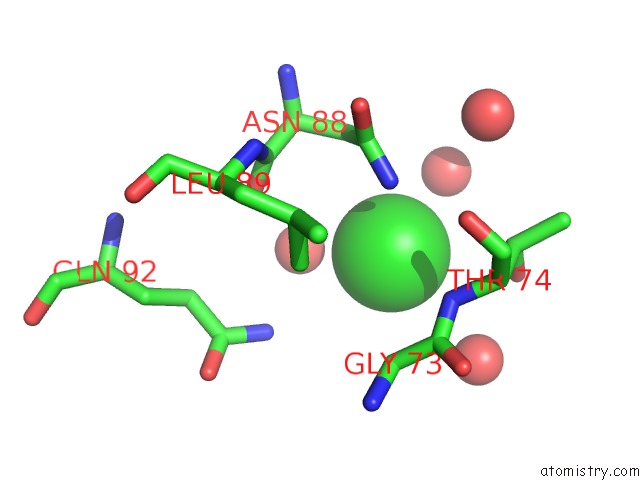
Mono view
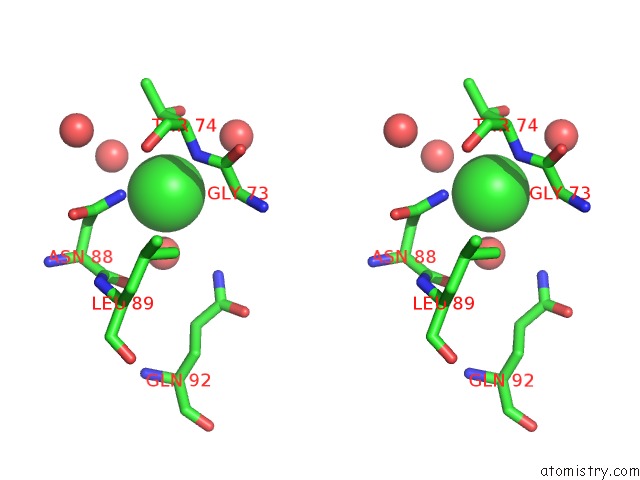
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Hiv-1 Wild Type Protease with Grl-0518A , An Isophthalamide-Derived P2-P3 Ligand with the Para-Hydoxymethyl Sulfonamide Isostere As the P2' Group within 5.0Å range:
|
Reference:
A.K.Ghosh,
M.Brindisi,
P.R.Nyalapatla,
J.Takayama,
J.R.Ella-Menye,
S.Yashchuk,
J.Agniswamy,
Y.F.Wang,
M.Aoki,
M.Amano,
I.T.Weber,
H.Mitsuya.
Design of Novel Hiv-1 Protease Inhibitors Incorporating Isophthalamide-Derived P2-P3 Ligands: Synthesis, Biological Evaluation and X-Ray Structural Studies of Inhibitor-Hiv-1 Protease Complex. Bioorg. Med. Chem. V. 25 5114 2017.
ISSN: ESSN 1464-3391
PubMed: 28434781
DOI: 10.1016/J.BMC.2017.04.005
Page generated: Sat Jul 12 09:27:13 2025
ISSN: ESSN 1464-3391
PubMed: 28434781
DOI: 10.1016/J.BMC.2017.04.005
Last articles
K in 5EDUK in 5EEK
K in 5EC2
K in 5EE9
K in 5DKP
K in 5EC1
K in 5EBW
K in 5EBL
K in 5EBM
K in 5DOU