Chlorine »
PDB 5vcr-5vmq »
5vjq »
Chlorine in PDB 5vjq: Complex Between HYHEL10 Fab Fragment Heavy Chain Mutant (I29F, S52T, Y53F) and Pekin Duck Egg Lysozyme Isoform I (Del-I)
Enzymatic activity of Complex Between HYHEL10 Fab Fragment Heavy Chain Mutant (I29F, S52T, Y53F) and Pekin Duck Egg Lysozyme Isoform I (Del-I)
All present enzymatic activity of Complex Between HYHEL10 Fab Fragment Heavy Chain Mutant (I29F, S52T, Y53F) and Pekin Duck Egg Lysozyme Isoform I (Del-I):
3.2.1.18;
3.2.1.18;
Protein crystallography data
The structure of Complex Between HYHEL10 Fab Fragment Heavy Chain Mutant (I29F, S52T, Y53F) and Pekin Duck Egg Lysozyme Isoform I (Del-I), PDB code: 5vjq
was solved by
D.B.Langley,
D.Christ,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 49.32 / 1.90 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 88.650, 132.660, 199.110, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 19.3 / 23.5 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Complex Between HYHEL10 Fab Fragment Heavy Chain Mutant (I29F, S52T, Y53F) and Pekin Duck Egg Lysozyme Isoform I (Del-I)
(pdb code 5vjq). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 4 binding sites of Chlorine where determined in the Complex Between HYHEL10 Fab Fragment Heavy Chain Mutant (I29F, S52T, Y53F) and Pekin Duck Egg Lysozyme Isoform I (Del-I), PDB code: 5vjq:
Jump to Chlorine binding site number: 1; 2; 3; 4;
In total 4 binding sites of Chlorine where determined in the Complex Between HYHEL10 Fab Fragment Heavy Chain Mutant (I29F, S52T, Y53F) and Pekin Duck Egg Lysozyme Isoform I (Del-I), PDB code: 5vjq:
Jump to Chlorine binding site number: 1; 2; 3; 4;
Chlorine binding site 1 out of 4 in 5vjq
Go back to
Chlorine binding site 1 out
of 4 in the Complex Between HYHEL10 Fab Fragment Heavy Chain Mutant (I29F, S52T, Y53F) and Pekin Duck Egg Lysozyme Isoform I (Del-I)
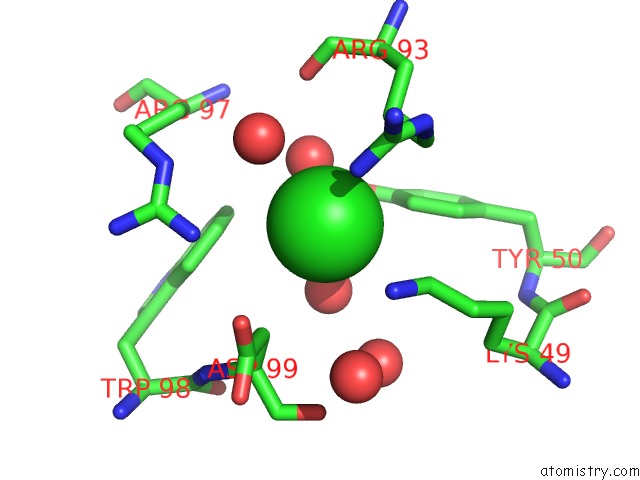
Mono view
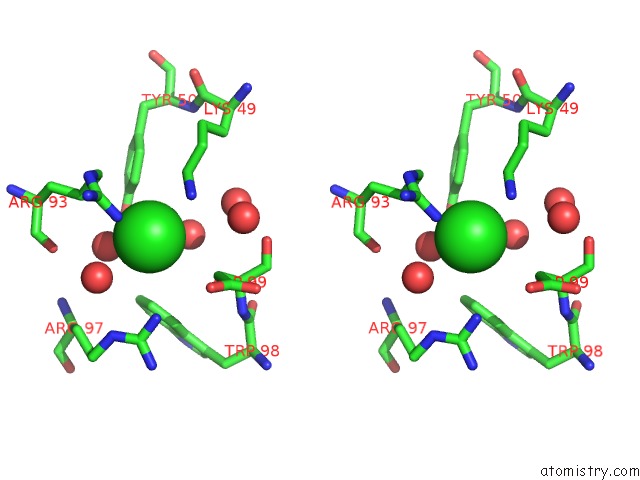
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Complex Between HYHEL10 Fab Fragment Heavy Chain Mutant (I29F, S52T, Y53F) and Pekin Duck Egg Lysozyme Isoform I (Del-I) within 5.0Å range:
|
Chlorine binding site 2 out of 4 in 5vjq
Go back to
Chlorine binding site 2 out
of 4 in the Complex Between HYHEL10 Fab Fragment Heavy Chain Mutant (I29F, S52T, Y53F) and Pekin Duck Egg Lysozyme Isoform I (Del-I)
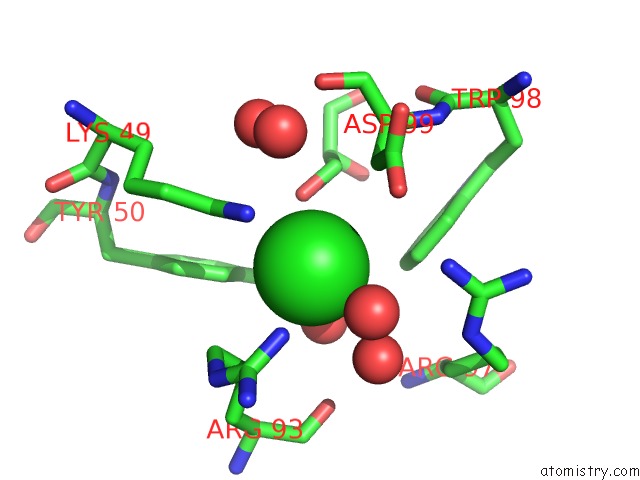
Mono view
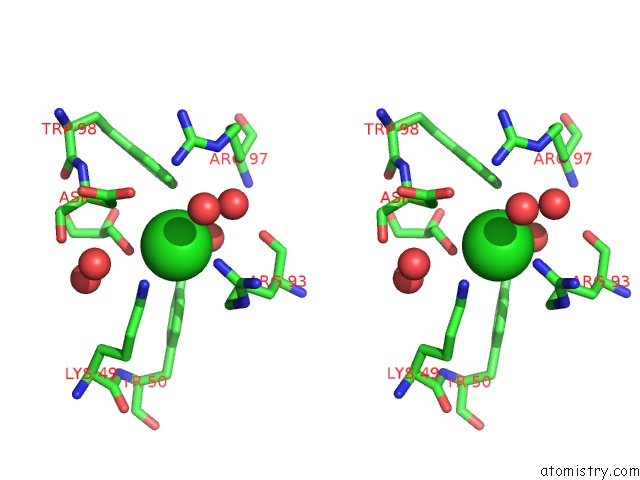
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Complex Between HYHEL10 Fab Fragment Heavy Chain Mutant (I29F, S52T, Y53F) and Pekin Duck Egg Lysozyme Isoform I (Del-I) within 5.0Å range:
|
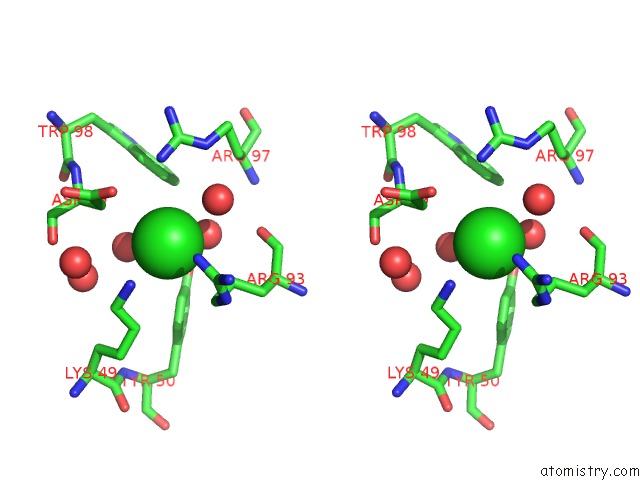
Chlorine binding site 3 out of 4 in 5vjq
Go back to
Chlorine binding site 3 out
of 4 in the Complex Between HYHEL10 Fab Fragment Heavy Chain Mutant (I29F, S52T, Y53F) and Pekin Duck Egg Lysozyme Isoform I (Del-I)
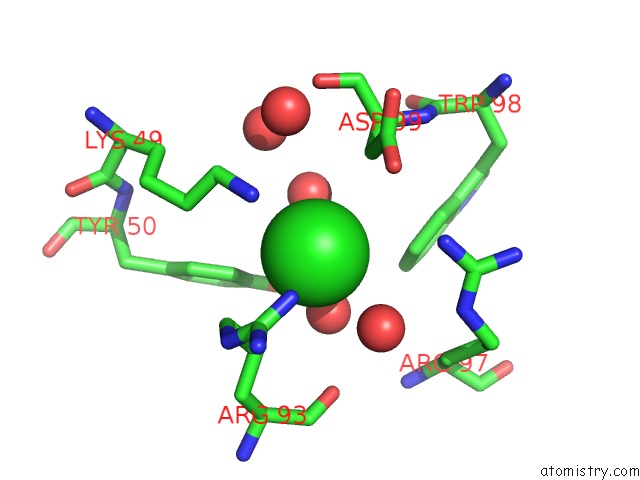
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Complex Between HYHEL10 Fab Fragment Heavy Chain Mutant (I29F, S52T, Y53F) and Pekin Duck Egg Lysozyme Isoform I (Del-I) within 5.0Å range:
|
Chlorine binding site 4 out of 4 in 5vjq
Go back to
Chlorine binding site 4 out
of 4 in the Complex Between HYHEL10 Fab Fragment Heavy Chain Mutant (I29F, S52T, Y53F) and Pekin Duck Egg Lysozyme Isoform I (Del-I)
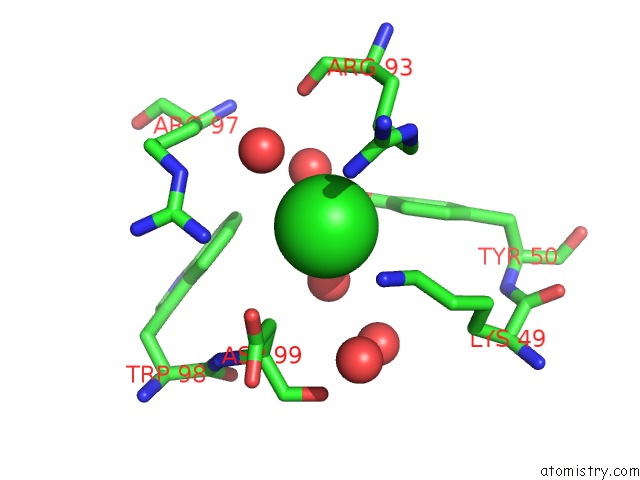
Mono view
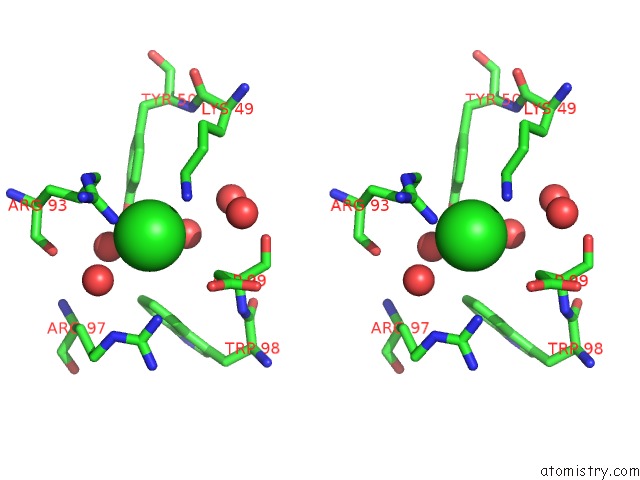
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of Complex Between HYHEL10 Fab Fragment Heavy Chain Mutant (I29F, S52T, Y53F) and Pekin Duck Egg Lysozyme Isoform I (Del-I) within 5.0Å range:
|
Reference:
D.L.Burnett,
D.B.Langley,
P.Schofield,
J.R.Hermes,
T.D.Chan,
J.Jackson,
K.Bourne,
J.H.Reed,
K.Patterson,
B.T.Porebski,
R.Brink,
D.Christ,
C.C.Goodnow.
Germinal Center Antibody Mutation Trajectories Are Determined By Rapid Self/Foreign Discrimination. Science V. 360 223 2018.
ISSN: ESSN 1095-9203
PubMed: 29650674
DOI: 10.1126/SCIENCE.AAO3859
Page generated: Sat Jul 12 09:54:13 2025
ISSN: ESSN 1095-9203
PubMed: 29650674
DOI: 10.1126/SCIENCE.AAO3859
Last articles
K in 9F90K in 9ES6
K in 9EWD
K in 9ETN
K in 9ESI
K in 9ESH
K in 9ES4
K in 9ES5
K in 9ES2
K in 9ES0