Chlorine »
PDB 6bsg-6bzh »
6bvf »
Chlorine in PDB 6bvf: Cryo-Em Structure of Hepatitis B Virus T=4 Capsid in Complex with the Fluorescent Allosteric Modulator Hap-Tamra
Other elements in 6bvf:
The structure of Cryo-Em Structure of Hepatitis B Virus T=4 Capsid in Complex with the Fluorescent Allosteric Modulator Hap-Tamra also contains other interesting chemical elements:
| Fluorine | (F) | 2 atoms |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Cryo-Em Structure of Hepatitis B Virus T=4 Capsid in Complex with the Fluorescent Allosteric Modulator Hap-Tamra
(pdb code 6bvf). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 2 binding sites of Chlorine where determined in the Cryo-Em Structure of Hepatitis B Virus T=4 Capsid in Complex with the Fluorescent Allosteric Modulator Hap-Tamra, PDB code: 6bvf:
Jump to Chlorine binding site number: 1; 2;
In total 2 binding sites of Chlorine where determined in the Cryo-Em Structure of Hepatitis B Virus T=4 Capsid in Complex with the Fluorescent Allosteric Modulator Hap-Tamra, PDB code: 6bvf:
Jump to Chlorine binding site number: 1; 2;
Chlorine binding site 1 out of 2 in 6bvf
Go back to
Chlorine binding site 1 out
of 2 in the Cryo-Em Structure of Hepatitis B Virus T=4 Capsid in Complex with the Fluorescent Allosteric Modulator Hap-Tamra
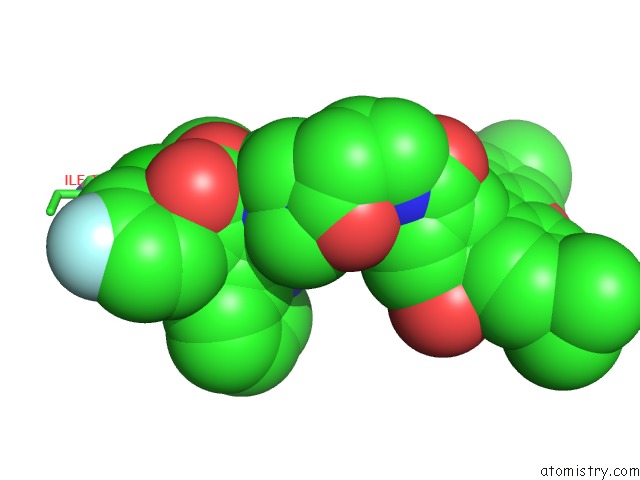
Mono view
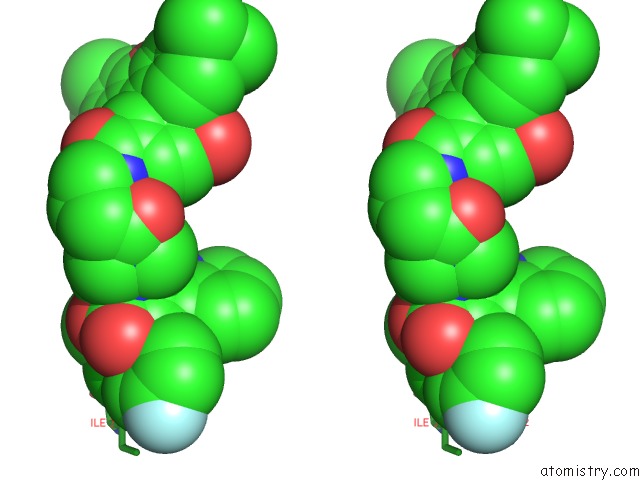
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Cryo-Em Structure of Hepatitis B Virus T=4 Capsid in Complex with the Fluorescent Allosteric Modulator Hap-Tamra within 5.0Å range:
|
Chlorine binding site 2 out of 2 in 6bvf
Go back to
Chlorine binding site 2 out
of 2 in the Cryo-Em Structure of Hepatitis B Virus T=4 Capsid in Complex with the Fluorescent Allosteric Modulator Hap-Tamra
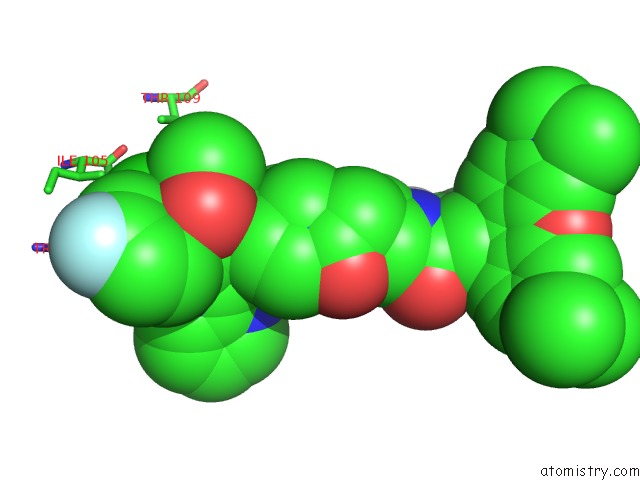
Mono view
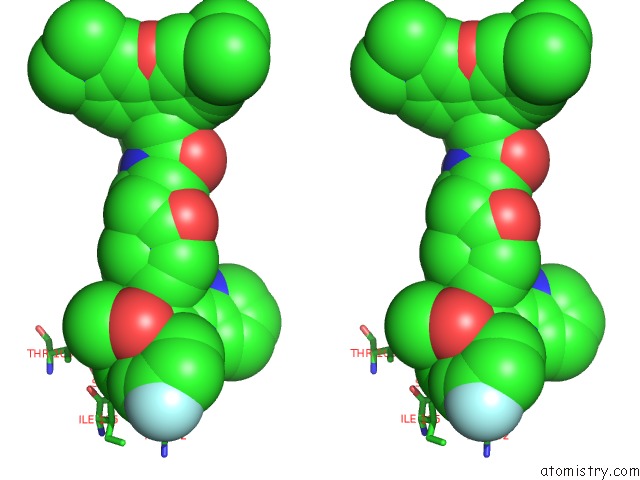
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Cryo-Em Structure of Hepatitis B Virus T=4 Capsid in Complex with the Fluorescent Allosteric Modulator Hap-Tamra within 5.0Å range:
|
Reference:
C.J.Schlicksup,
J.C.Wang,
S.Francis,
B.Venkatakrishnan,
W.W.Turner,
M.Vannieuwenhze,
A.Zlotnick.
Hepatitis B Virus Core Protein Allosteric Modulators Can Distort and Disrupt Intact Capsids. Elife V. 7 2018.
ISSN: ESSN 2050-084X
PubMed: 29377794
DOI: 10.7554/ELIFE.31473
Page generated: Sat Jul 12 12:07:37 2025
ISSN: ESSN 2050-084X
PubMed: 29377794
DOI: 10.7554/ELIFE.31473
Last articles
Mo in 3ML1Mo in 3K6X
Mo in 3L4P
Mo in 3K1A
Mo in 3K6W
Mo in 3GZG
Mo in 3HRD
Mo in 3IR5
Mo in 3IR7
Mo in 3HBQ