Chlorine »
PDB 6dox-6dy2 »
6dq8 »
Chlorine in PDB 6dq8: Linked KDM5A Jmj Domain Bound to the Inhibitor N49 I.E. 2-((2- Chlorophenyl)(2-(1-Methylpyrrolidin-2-Yl)Ethoxy)Methyl)Thieno[3,2- B]Pyridine-7-Carboxylic Acid
Protein crystallography data
The structure of Linked KDM5A Jmj Domain Bound to the Inhibitor N49 I.E. 2-((2- Chlorophenyl)(2-(1-Methylpyrrolidin-2-Yl)Ethoxy)Methyl)Thieno[3,2- B]Pyridine-7-Carboxylic Acid, PDB code: 6dq8
was solved by
J.R.Horton,
X.Cheng,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 33.01 / 1.46 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 117.253, 61.754, 46.718, 90.00, 92.33, 90.00 |
| R / Rfree (%) | 18.6 / 21 |
Other elements in 6dq8:
The structure of Linked KDM5A Jmj Domain Bound to the Inhibitor N49 I.E. 2-((2- Chlorophenyl)(2-(1-Methylpyrrolidin-2-Yl)Ethoxy)Methyl)Thieno[3,2- B]Pyridine-7-Carboxylic Acid also contains other interesting chemical elements:
| Manganese | (Mn) | 1 atom |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Linked KDM5A Jmj Domain Bound to the Inhibitor N49 I.E. 2-((2- Chlorophenyl)(2-(1-Methylpyrrolidin-2-Yl)Ethoxy)Methyl)Thieno[3,2- B]Pyridine-7-Carboxylic Acid
(pdb code 6dq8). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 2 binding sites of Chlorine where determined in the Linked KDM5A Jmj Domain Bound to the Inhibitor N49 I.E. 2-((2- Chlorophenyl)(2-(1-Methylpyrrolidin-2-Yl)Ethoxy)Methyl)Thieno[3,2- B]Pyridine-7-Carboxylic Acid, PDB code: 6dq8:
Jump to Chlorine binding site number: 1; 2;
In total 2 binding sites of Chlorine where determined in the Linked KDM5A Jmj Domain Bound to the Inhibitor N49 I.E. 2-((2- Chlorophenyl)(2-(1-Methylpyrrolidin-2-Yl)Ethoxy)Methyl)Thieno[3,2- B]Pyridine-7-Carboxylic Acid, PDB code: 6dq8:
Jump to Chlorine binding site number: 1; 2;
Chlorine binding site 1 out of 2 in 6dq8
Go back to
Chlorine binding site 1 out
of 2 in the Linked KDM5A Jmj Domain Bound to the Inhibitor N49 I.E. 2-((2- Chlorophenyl)(2-(1-Methylpyrrolidin-2-Yl)Ethoxy)Methyl)Thieno[3,2- B]Pyridine-7-Carboxylic Acid
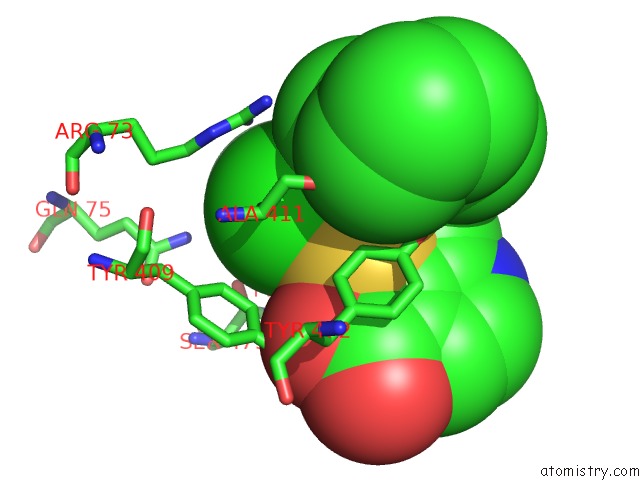
Mono view
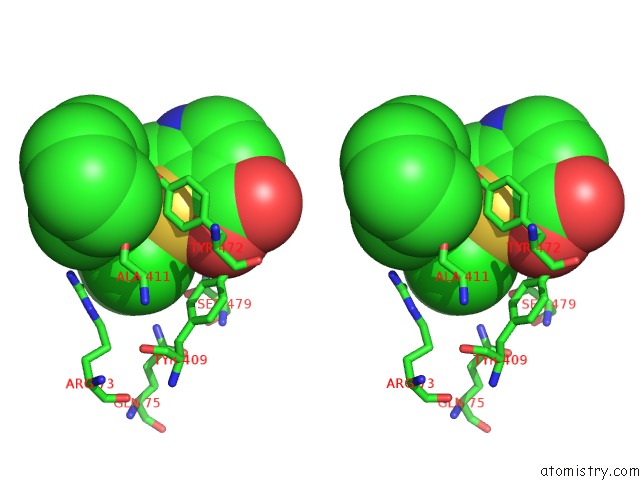
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Linked KDM5A Jmj Domain Bound to the Inhibitor N49 I.E. 2-((2- Chlorophenyl)(2-(1-Methylpyrrolidin-2-Yl)Ethoxy)Methyl)Thieno[3,2- B]Pyridine-7-Carboxylic Acid within 5.0Å range:
|
Chlorine binding site 2 out of 2 in 6dq8
Go back to
Chlorine binding site 2 out
of 2 in the Linked KDM5A Jmj Domain Bound to the Inhibitor N49 I.E. 2-((2- Chlorophenyl)(2-(1-Methylpyrrolidin-2-Yl)Ethoxy)Methyl)Thieno[3,2- B]Pyridine-7-Carboxylic Acid
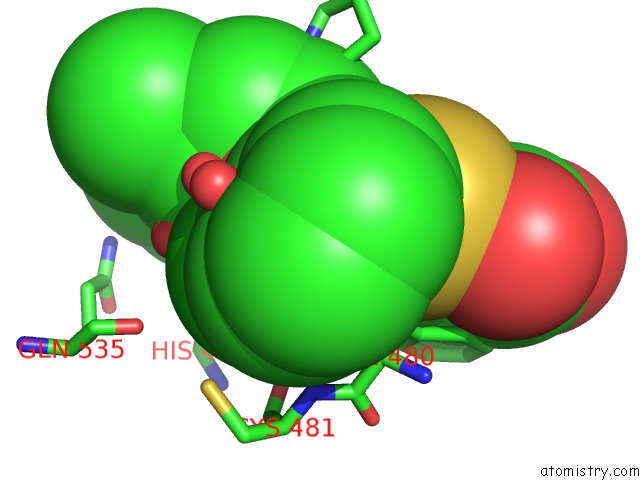
Mono view
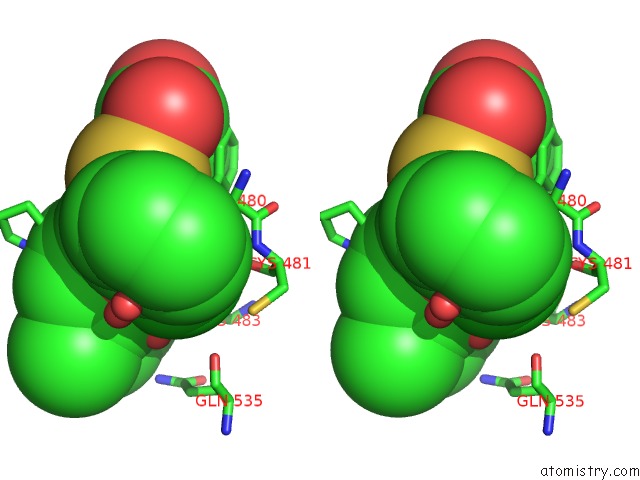
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Linked KDM5A Jmj Domain Bound to the Inhibitor N49 I.E. 2-((2- Chlorophenyl)(2-(1-Methylpyrrolidin-2-Yl)Ethoxy)Methyl)Thieno[3,2- B]Pyridine-7-Carboxylic Acid within 5.0Å range:
|
Reference:
J.R.Horton,
C.B.Woodcock,
Q.Chen,
X.Liu,
X.Zhang,
J.Shanks,
G.Rai,
B.T.Mott,
D.J.Jansen,
S.C.Kales,
M.J.Henderson,
M.Cyr,
K.Pohida,
X.Hu,
P.Shah,
X.Xu,
A.Jadhav,
D.J.Maloney,
M.D.Hall,
A.Simeonov,
H.Fu,
P.M.Vertino,
X.Cheng.
Structure-Based Engineering of Irreversible Inhibitors Against Histone Lysine Demethylase KDM5A. J. Med. Chem. V. 61 10588 2018.
ISSN: ISSN 1520-4804
PubMed: 30392349
DOI: 10.1021/ACS.JMEDCHEM.8B01219
Page generated: Sat Jul 12 13:00:17 2025
ISSN: ISSN 1520-4804
PubMed: 30392349
DOI: 10.1021/ACS.JMEDCHEM.8B01219
Last articles
K in 8OFDK in 8OEO
K in 8OED
K in 8OEH
K in 8K1Z
K in 8K1U
K in 8K1V
K in 8K7W
K in 8K1T
K in 8K1S