Chlorine »
PDB 6e7r-6el4 »
6eg3 »
Chlorine in PDB 6eg3: Crystal Structure of Human Brm in Complex with Compound 15
Protein crystallography data
The structure of Crystal Structure of Human Brm in Complex with Compound 15, PDB code: 6eg3
was solved by
X.Zhu,
R.Kulathila,
T.Hu,
X.Xie,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 96.50 / 2.84 |
| Space group | P 42 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 136.470, 136.470, 121.370, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 22.9 / 26.7 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Crystal Structure of Human Brm in Complex with Compound 15
(pdb code 6eg3). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total only one binding site of Chlorine was determined in the Crystal Structure of Human Brm in Complex with Compound 15, PDB code: 6eg3:
In total only one binding site of Chlorine was determined in the Crystal Structure of Human Brm in Complex with Compound 15, PDB code: 6eg3:
Chlorine binding site 1 out of 1 in 6eg3
Go back to
Chlorine binding site 1 out
of 1 in the Crystal Structure of Human Brm in Complex with Compound 15
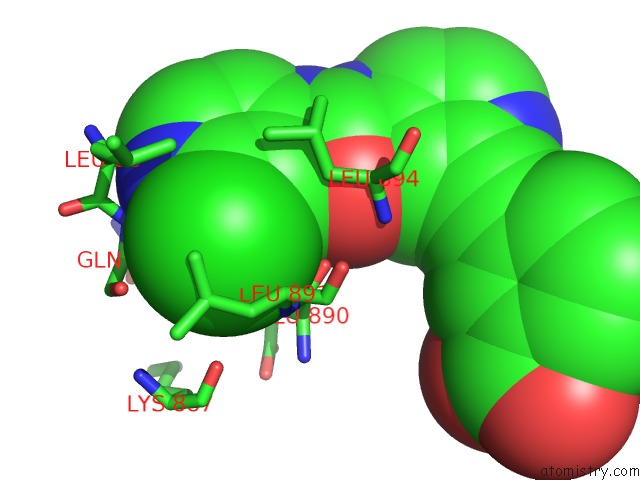
Mono view
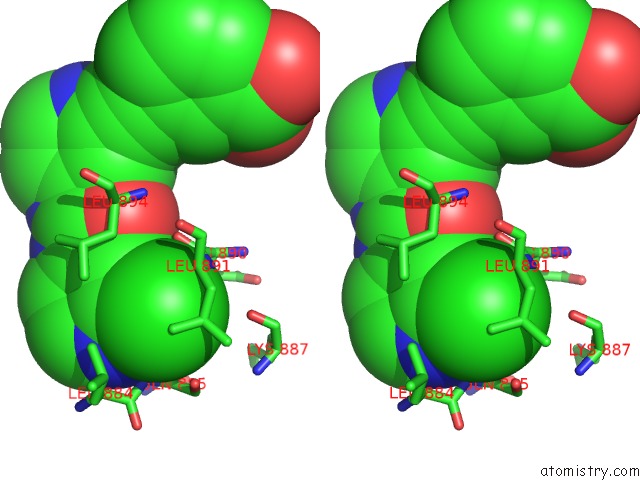
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Crystal Structure of Human Brm in Complex with Compound 15 within 5.0Å range:
|
Reference:
J.P.N.Papillon,
K.Nakajima,
C.D.Adair,
J.Hempel,
A.O.Jouk,
R.G.Karki,
S.Mathieu,
H.Mobitz,
R.Ntaganda,
T.Smith,
M.Visser,
S.E.Hill,
F.K.Hurtado,
G.Chenail,
H.C.Bhang,
A.Bric,
K.Xiang,
G.Bushold,
T.Gilbert,
A.Vattay,
J.Dooley,
E.A.Costa,
I.Park,
A.Li,
D.Farley,
E.Lounkine,
Q.K.Yue,
X.Xie,
X.Zhu,
R.Kulathila,
D.King,
T.Hu,
K.Vulic,
J.Cantwell,
C.Luu,
Z.Jagani.
Discovery of Orally Active Inhibitors of Brahma Homolog (Brm)/SMARCA2 Atpase Activity For the Treatment of Brahma Related Gene 1 (BRG1)/SMARCA4-Mutant Cancers. J. Med. Chem. V. 61 10155 2018.
ISSN: ISSN 1520-4804
PubMed: 30339381
DOI: 10.1021/ACS.JMEDCHEM.8B01318
Page generated: Sat Jul 12 13:25:37 2025
ISSN: ISSN 1520-4804
PubMed: 30339381
DOI: 10.1021/ACS.JMEDCHEM.8B01318
Last articles
Mn in 8V7FMn in 8UWB
Mn in 8UW0
Mn in 8UYR
Mn in 8UZ8
Mn in 8UX7
Mn in 8UN2
Mn in 8UJG
Mn in 8UJF
Mn in 8UMC