Chlorine »
PDB 6ero-6ezp »
6eup »
Chlorine in PDB 6eup: Crystal Structure of Neisseria Meningitidis Nada Variant 3 Double Mutant A33I-I38L
Protein crystallography data
The structure of Crystal Structure of Neisseria Meningitidis Nada Variant 3 Double Mutant A33I-I38L, PDB code: 6eup
was solved by
L.Dello Iacono,
A.Liguori,
E.Malito,
M.J.Bottomley,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 63.65 / 2.65 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 68.919, 39.750, 255.630, 90.00, 95.13, 90.00 |
| R / Rfree (%) | 19.8 / 23.6 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Crystal Structure of Neisseria Meningitidis Nada Variant 3 Double Mutant A33I-I38L
(pdb code 6eup). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total only one binding site of Chlorine was determined in the Crystal Structure of Neisseria Meningitidis Nada Variant 3 Double Mutant A33I-I38L, PDB code: 6eup:
In total only one binding site of Chlorine was determined in the Crystal Structure of Neisseria Meningitidis Nada Variant 3 Double Mutant A33I-I38L, PDB code: 6eup:
Chlorine binding site 1 out of 1 in 6eup
Go back to
Chlorine binding site 1 out
of 1 in the Crystal Structure of Neisseria Meningitidis Nada Variant 3 Double Mutant A33I-I38L
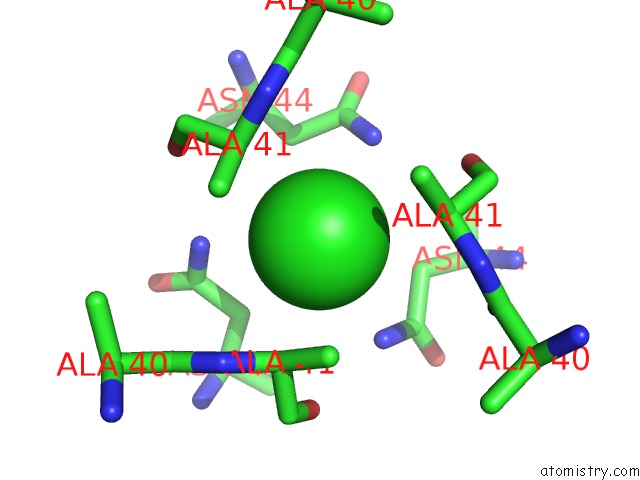
Mono view
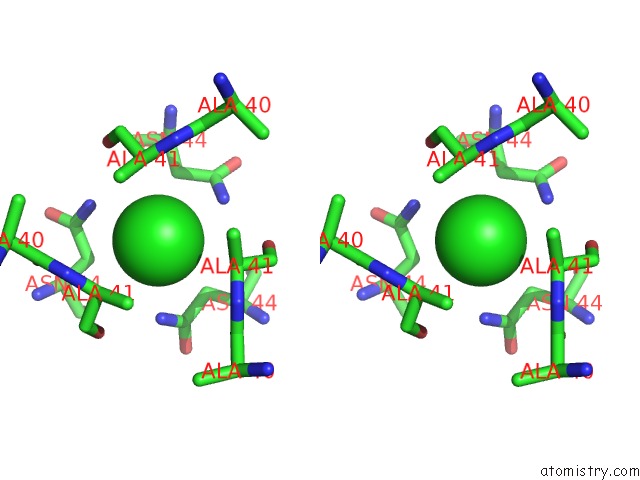
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Crystal Structure of Neisseria Meningitidis Nada Variant 3 Double Mutant A33I-I38L within 5.0Å range:
|
Reference:
A.Liguori,
L.Dello Iacono,
G.Maruggi,
B.Benucci,
M.Merola,
P.Lo Surdo,
J.Lopez-Sagaseta,
M.Pizza,
E.Malito,
M.J.Bottomley.
NADA3 Structures Reveal Undecad Coiled Coils and LOX1 Binding Regions Competed By Meningococcus B Vaccine-Elicited Human Antibodies. Mbio V. 9 2018.
ISSN: ESSN 2150-7511
PubMed: 30327444
DOI: 10.1128/MBIO.01914-18
Page generated: Sat Jul 12 13:37:46 2025
ISSN: ESSN 2150-7511
PubMed: 30327444
DOI: 10.1128/MBIO.01914-18
Last articles
Mg in 9GUPMg in 9GUR
Mg in 9GRE
Mg in 9GTK
Mg in 9GU5
Mg in 9GOB
Mg in 9GO5
Mg in 9GMZ
Mg in 9GQO
Mg in 9GMX