Chlorine »
PDB 6ggr-6gp4 »
6gob »
Chlorine in PDB 6gob: X-Ray Structure of the Adduct Formed Upon Reaction of Lysozyme with A Pd(II) Complex Bearing N,N-Pyridylbenzimidazole Derivative with An Alkylated Sulphonate Side Chain
Enzymatic activity of X-Ray Structure of the Adduct Formed Upon Reaction of Lysozyme with A Pd(II) Complex Bearing N,N-Pyridylbenzimidazole Derivative with An Alkylated Sulphonate Side Chain
All present enzymatic activity of X-Ray Structure of the Adduct Formed Upon Reaction of Lysozyme with A Pd(II) Complex Bearing N,N-Pyridylbenzimidazole Derivative with An Alkylated Sulphonate Side Chain:
3.2.1.17;
3.2.1.17;
Protein crystallography data
The structure of X-Ray Structure of the Adduct Formed Upon Reaction of Lysozyme with A Pd(II) Complex Bearing N,N-Pyridylbenzimidazole Derivative with An Alkylated Sulphonate Side Chain, PDB code: 6gob
was solved by
A.Merlino,
G.Ferraro,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 56.21 / 1.96 |
| Space group | P 43 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 79.487, 79.487, 37.309, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 16.9 / 23.1 |
Other elements in 6gob:
The structure of X-Ray Structure of the Adduct Formed Upon Reaction of Lysozyme with A Pd(II) Complex Bearing N,N-Pyridylbenzimidazole Derivative with An Alkylated Sulphonate Side Chain also contains other interesting chemical elements:
| Palladium | (Pd) | 7 atoms |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the X-Ray Structure of the Adduct Formed Upon Reaction of Lysozyme with A Pd(II) Complex Bearing N,N-Pyridylbenzimidazole Derivative with An Alkylated Sulphonate Side Chain
(pdb code 6gob). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 8 binding sites of Chlorine where determined in the X-Ray Structure of the Adduct Formed Upon Reaction of Lysozyme with A Pd(II) Complex Bearing N,N-Pyridylbenzimidazole Derivative with An Alkylated Sulphonate Side Chain, PDB code: 6gob:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5; 6; 7; 8;
In total 8 binding sites of Chlorine where determined in the X-Ray Structure of the Adduct Formed Upon Reaction of Lysozyme with A Pd(II) Complex Bearing N,N-Pyridylbenzimidazole Derivative with An Alkylated Sulphonate Side Chain, PDB code: 6gob:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5; 6; 7; 8;
Chlorine binding site 1 out of 8 in 6gob
Go back to
Chlorine binding site 1 out
of 8 in the X-Ray Structure of the Adduct Formed Upon Reaction of Lysozyme with A Pd(II) Complex Bearing N,N-Pyridylbenzimidazole Derivative with An Alkylated Sulphonate Side Chain
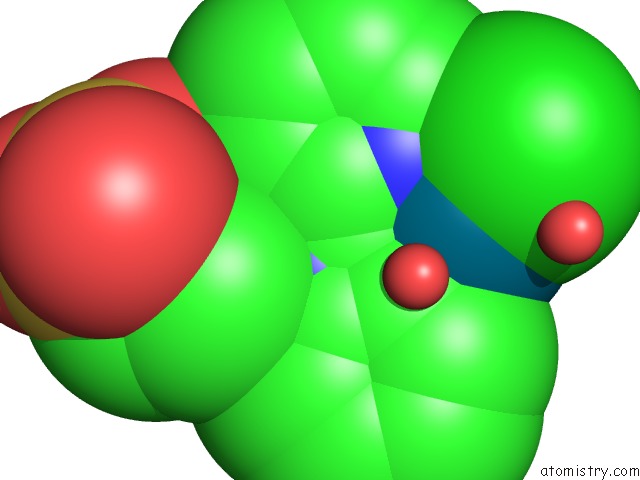
Mono view
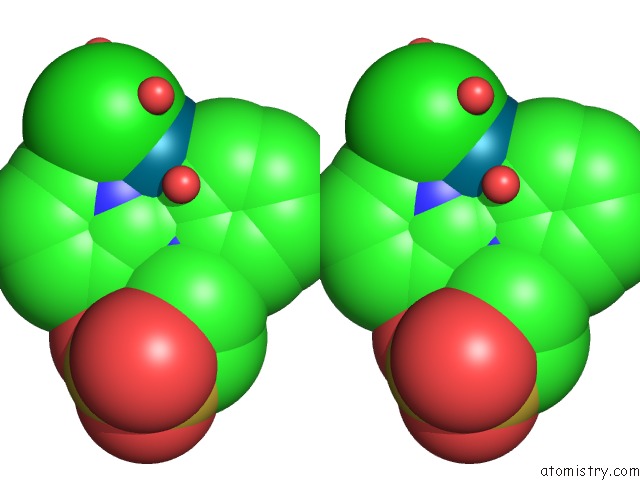
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of X-Ray Structure of the Adduct Formed Upon Reaction of Lysozyme with A Pd(II) Complex Bearing N,N-Pyridylbenzimidazole Derivative with An Alkylated Sulphonate Side Chain within 5.0Å range:
|
Chlorine binding site 2 out of 8 in 6gob
Go back to
Chlorine binding site 2 out
of 8 in the X-Ray Structure of the Adduct Formed Upon Reaction of Lysozyme with A Pd(II) Complex Bearing N,N-Pyridylbenzimidazole Derivative with An Alkylated Sulphonate Side Chain
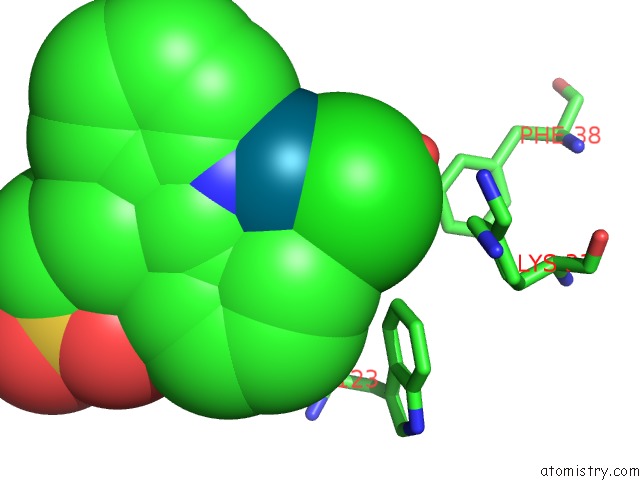
Mono view
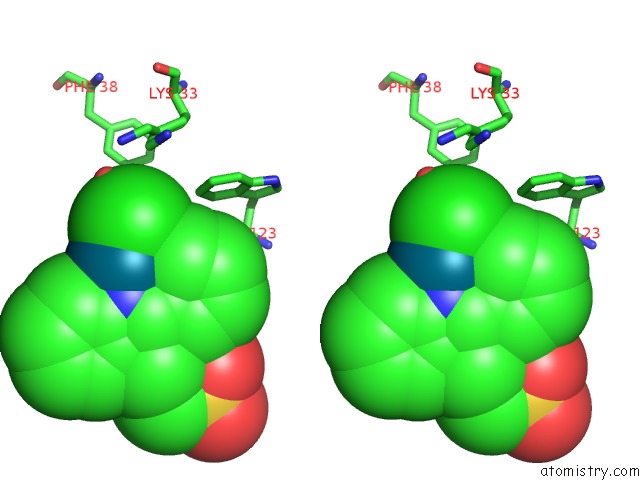
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of X-Ray Structure of the Adduct Formed Upon Reaction of Lysozyme with A Pd(II) Complex Bearing N,N-Pyridylbenzimidazole Derivative with An Alkylated Sulphonate Side Chain within 5.0Å range:
|
Chlorine binding site 3 out of 8 in 6gob
Go back to
Chlorine binding site 3 out
of 8 in the X-Ray Structure of the Adduct Formed Upon Reaction of Lysozyme with A Pd(II) Complex Bearing N,N-Pyridylbenzimidazole Derivative with An Alkylated Sulphonate Side Chain
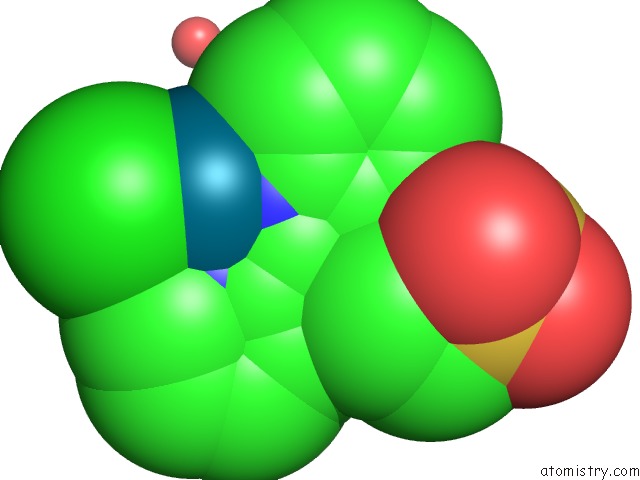
Mono view
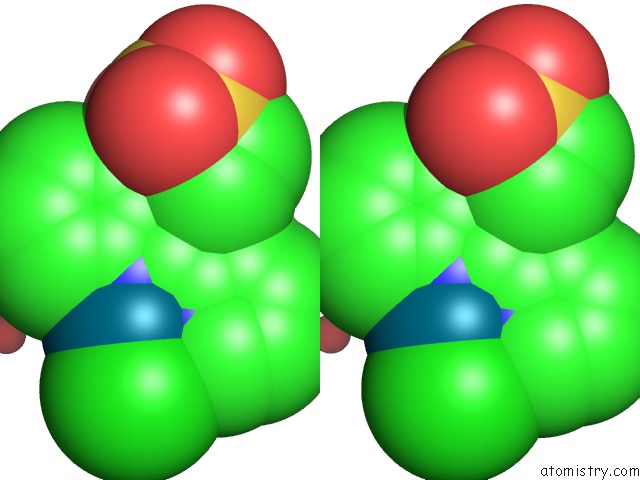
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of X-Ray Structure of the Adduct Formed Upon Reaction of Lysozyme with A Pd(II) Complex Bearing N,N-Pyridylbenzimidazole Derivative with An Alkylated Sulphonate Side Chain within 5.0Å range:
|
Chlorine binding site 4 out of 8 in 6gob
Go back to
Chlorine binding site 4 out
of 8 in the X-Ray Structure of the Adduct Formed Upon Reaction of Lysozyme with A Pd(II) Complex Bearing N,N-Pyridylbenzimidazole Derivative with An Alkylated Sulphonate Side Chain
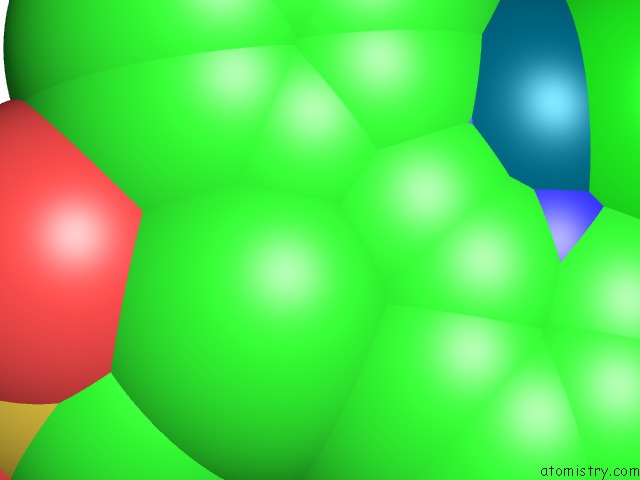
Mono view
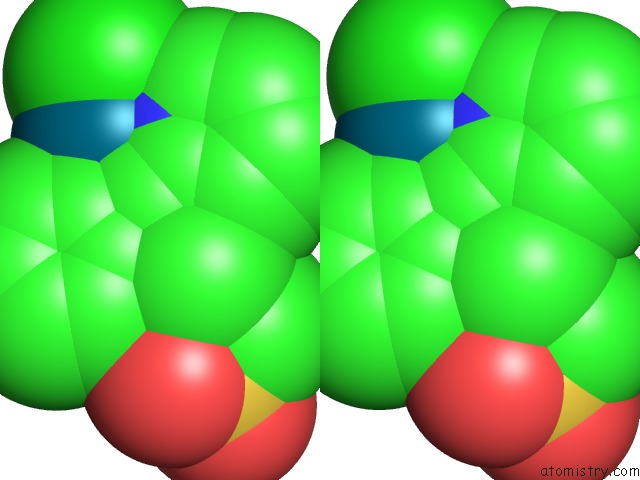
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of X-Ray Structure of the Adduct Formed Upon Reaction of Lysozyme with A Pd(II) Complex Bearing N,N-Pyridylbenzimidazole Derivative with An Alkylated Sulphonate Side Chain within 5.0Å range:
|
Chlorine binding site 5 out of 8 in 6gob
Go back to
Chlorine binding site 5 out
of 8 in the X-Ray Structure of the Adduct Formed Upon Reaction of Lysozyme with A Pd(II) Complex Bearing N,N-Pyridylbenzimidazole Derivative with An Alkylated Sulphonate Side Chain
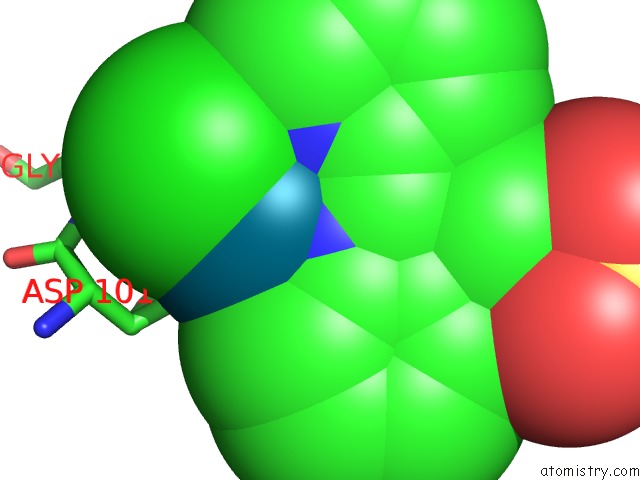
Mono view
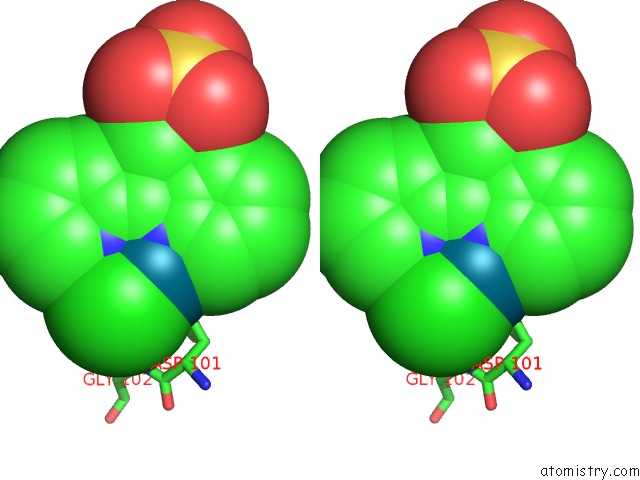
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 5 of X-Ray Structure of the Adduct Formed Upon Reaction of Lysozyme with A Pd(II) Complex Bearing N,N-Pyridylbenzimidazole Derivative with An Alkylated Sulphonate Side Chain within 5.0Å range:
|
Chlorine binding site 6 out of 8 in 6gob
Go back to
Chlorine binding site 6 out
of 8 in the X-Ray Structure of the Adduct Formed Upon Reaction of Lysozyme with A Pd(II) Complex Bearing N,N-Pyridylbenzimidazole Derivative with An Alkylated Sulphonate Side Chain
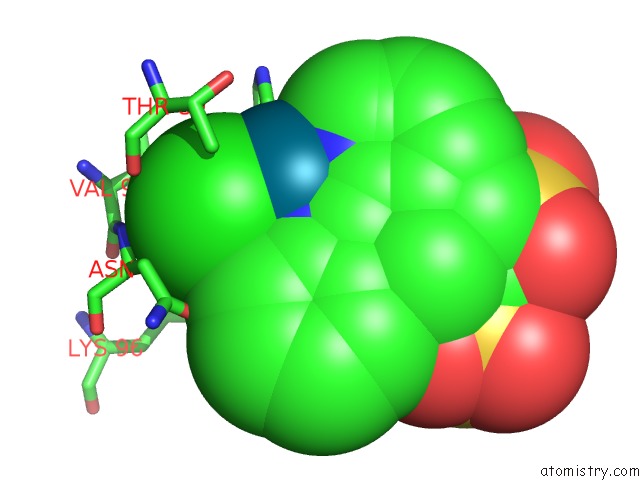
Mono view
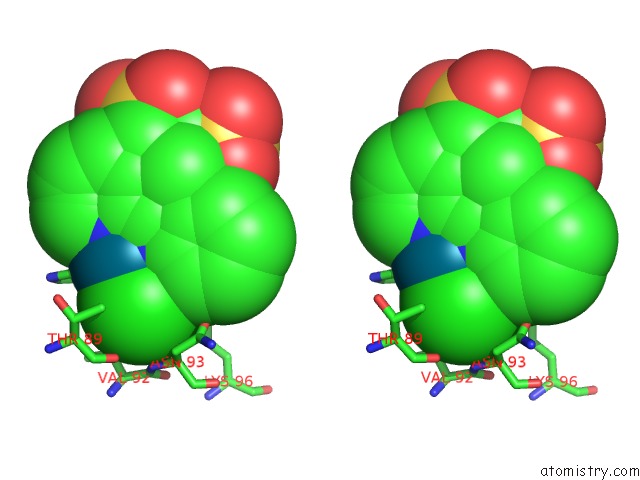
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 6 of X-Ray Structure of the Adduct Formed Upon Reaction of Lysozyme with A Pd(II) Complex Bearing N,N-Pyridylbenzimidazole Derivative with An Alkylated Sulphonate Side Chain within 5.0Å range:
|
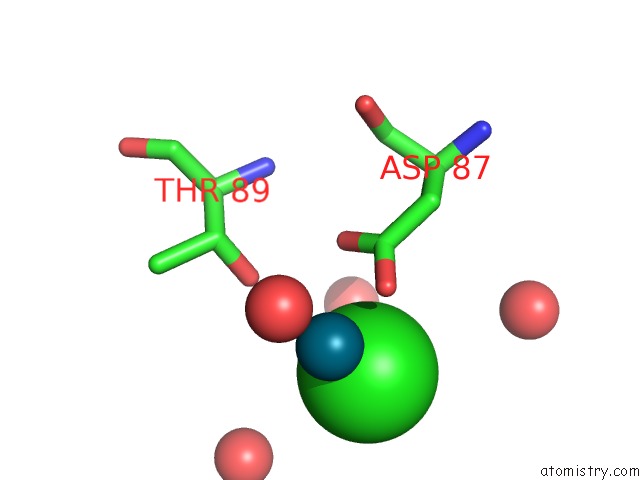
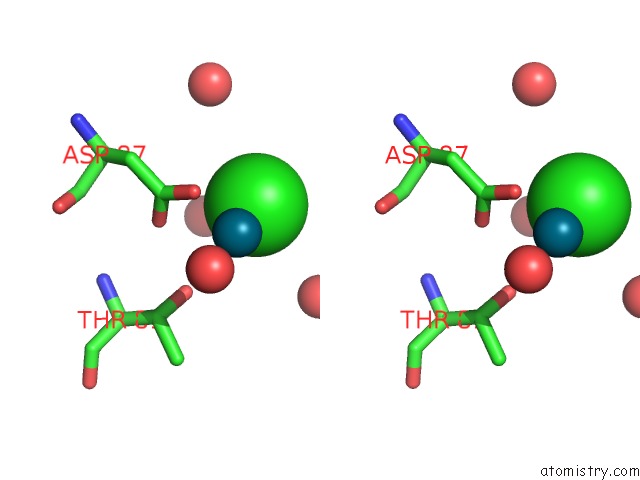
Chlorine binding site 7 out of 8 in 6gob
Go back to
Chlorine binding site 7 out
of 8 in the X-Ray Structure of the Adduct Formed Upon Reaction of Lysozyme with A Pd(II) Complex Bearing N,N-Pyridylbenzimidazole Derivative with An Alkylated Sulphonate Side Chain
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 7 of X-Ray Structure of the Adduct Formed Upon Reaction of Lysozyme with A Pd(II) Complex Bearing N,N-Pyridylbenzimidazole Derivative with An Alkylated Sulphonate Side Chain within 5.0Å range:
|
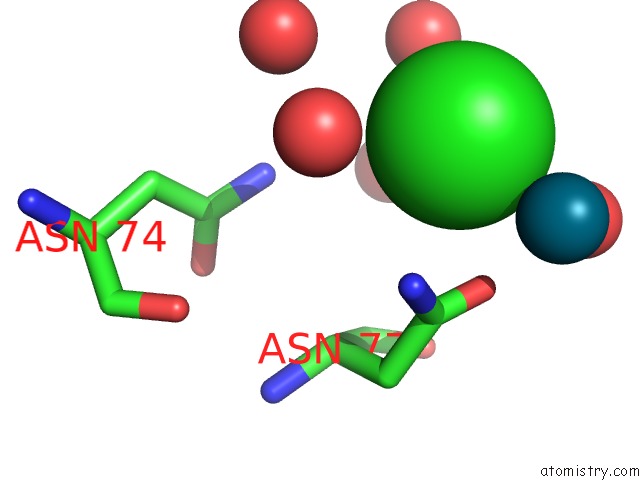
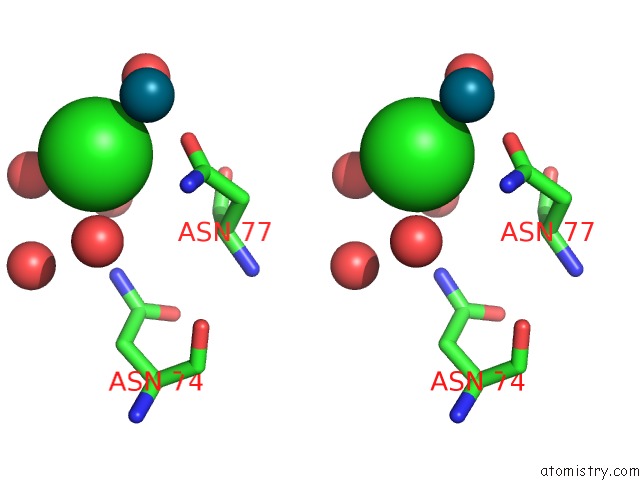
Chlorine binding site 8 out of 8 in 6gob
Go back to
Chlorine binding site 8 out
of 8 in the X-Ray Structure of the Adduct Formed Upon Reaction of Lysozyme with A Pd(II) Complex Bearing N,N-Pyridylbenzimidazole Derivative with An Alkylated Sulphonate Side Chain
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 8 of X-Ray Structure of the Adduct Formed Upon Reaction of Lysozyme with A Pd(II) Complex Bearing N,N-Pyridylbenzimidazole Derivative with An Alkylated Sulphonate Side Chain within 5.0Å range:
|
Reference:
G.Ferraro,
A.M.Mansour,
A.Merlino.
Exploring the Interactions Between Model Proteins and Pd(II) or Pt(II) Compounds Bearing Charged N,N-Pyridylbenzimidazole Bidentate Ligands By X-Ray Crystallography. Dalton Trans V. 47 10130 2018.
ISSN: ESSN 1477-9234
PubMed: 30004541
DOI: 10.1039/C8DT01663A
Page generated: Sat Jul 12 14:37:38 2025
ISSN: ESSN 1477-9234
PubMed: 30004541
DOI: 10.1039/C8DT01663A
Last articles
K in 8W9DK in 8W9C
K in 8W9E
K in 8W1Q
K in 8VQK
K in 8VZ7
K in 8VX0
K in 8VHH
K in 8VL8
K in 8VAZ