Chlorine »
PDB 6inc-6j1a »
6ipf »
Chlorine in PDB 6ipf: Post-Catalytic Complex of Human Dna Polymerase Mu with Templating Cytosine and Mn-8OXODGMP
Enzymatic activity of Post-Catalytic Complex of Human Dna Polymerase Mu with Templating Cytosine and Mn-8OXODGMP
All present enzymatic activity of Post-Catalytic Complex of Human Dna Polymerase Mu with Templating Cytosine and Mn-8OXODGMP:
2.7.7.7;
2.7.7.7;
Protein crystallography data
The structure of Post-Catalytic Complex of Human Dna Polymerase Mu with Templating Cytosine and Mn-8OXODGMP, PDB code: 6ipf
was solved by
Y.K.Chang,
W.J.Wu,
M.D.Tsai,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 30.00 / 1.77 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 59.778, 68.792, 109.620, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 18.8 / 21.8 |
Other elements in 6ipf:
The structure of Post-Catalytic Complex of Human Dna Polymerase Mu with Templating Cytosine and Mn-8OXODGMP also contains other interesting chemical elements:
| Manganese | (Mn) | 4 atoms |
| Sodium | (Na) | 2 atoms |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Post-Catalytic Complex of Human Dna Polymerase Mu with Templating Cytosine and Mn-8OXODGMP
(pdb code 6ipf). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total only one binding site of Chlorine was determined in the Post-Catalytic Complex of Human Dna Polymerase Mu with Templating Cytosine and Mn-8OXODGMP, PDB code: 6ipf:
In total only one binding site of Chlorine was determined in the Post-Catalytic Complex of Human Dna Polymerase Mu with Templating Cytosine and Mn-8OXODGMP, PDB code: 6ipf:
Chlorine binding site 1 out of 1 in 6ipf
Go back to
Chlorine binding site 1 out
of 1 in the Post-Catalytic Complex of Human Dna Polymerase Mu with Templating Cytosine and Mn-8OXODGMP
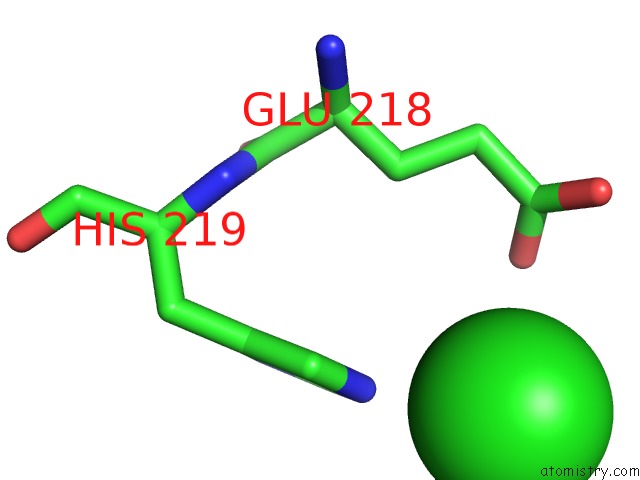
Mono view
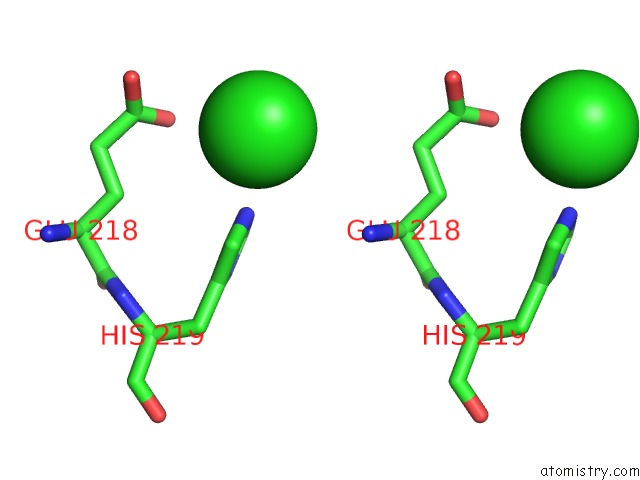
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Post-Catalytic Complex of Human Dna Polymerase Mu with Templating Cytosine and Mn-8OXODGMP within 5.0Å range:
|
Reference:
Y.K.Chang,
Y.P.Huang,
X.X.Liu,
T.P.Ko,
Y.Bessho,
Y.Kawano,
M.Maestre-Reyna,
W.J.Wu,
M.D.Tsai.
Human Dna Polymerase Mu Can Use A Noncanonical Mechanism For Multiple MN2+-Mediated Functions. J.Am.Chem.Soc. V. 141 8489 2019.
ISSN: ESSN 1520-5126
PubMed: 31067051
DOI: 10.1021/JACS.9B01741
Page generated: Sat Jul 12 15:47:55 2025
ISSN: ESSN 1520-5126
PubMed: 31067051
DOI: 10.1021/JACS.9B01741
Last articles
K in 7N9KK in 7MTX
K in 7MTU
K in 7N7C
K in 7N2M
K in 7MXO
K in 7MKT
K in 7MUB
K in 7MLW
K in 7MKY