Chlorine »
PDB 6j1b-6jlh »
6jke »
Chlorine in PDB 6jke: Discovery and the Crystal Structure of NS5 in Complex with the N- Terminal Bromodomain of BRD2.
Protein crystallography data
The structure of Discovery and the Crystal Structure of NS5 in Complex with the N- Terminal Bromodomain of BRD2., PDB code: 6jke
was solved by
B.Padmanabhan,
S.Mathur,
P.Deshmukh,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 67.67 / 1.50 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 114.368, 55.750, 67.850, 90.00, 94.16, 90.00 |
| R / Rfree (%) | 18.6 / 22.6 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Discovery and the Crystal Structure of NS5 in Complex with the N- Terminal Bromodomain of BRD2.
(pdb code 6jke). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 2 binding sites of Chlorine where determined in the Discovery and the Crystal Structure of NS5 in Complex with the N- Terminal Bromodomain of BRD2., PDB code: 6jke:
Jump to Chlorine binding site number: 1; 2;
In total 2 binding sites of Chlorine where determined in the Discovery and the Crystal Structure of NS5 in Complex with the N- Terminal Bromodomain of BRD2., PDB code: 6jke:
Jump to Chlorine binding site number: 1; 2;
Chlorine binding site 1 out of 2 in 6jke
Go back to
Chlorine binding site 1 out
of 2 in the Discovery and the Crystal Structure of NS5 in Complex with the N- Terminal Bromodomain of BRD2.
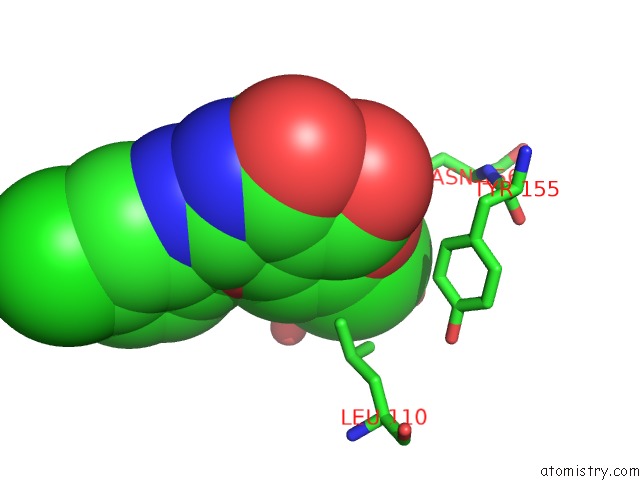
Mono view
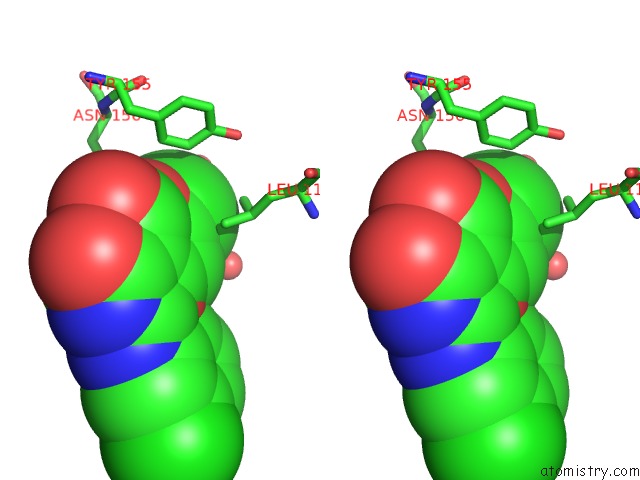
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Discovery and the Crystal Structure of NS5 in Complex with the N- Terminal Bromodomain of BRD2. within 5.0Å range:
|
Chlorine binding site 2 out of 2 in 6jke
Go back to
Chlorine binding site 2 out
of 2 in the Discovery and the Crystal Structure of NS5 in Complex with the N- Terminal Bromodomain of BRD2.
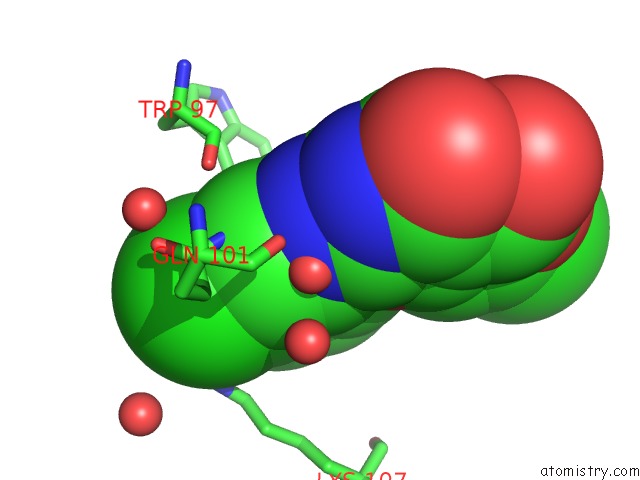
Mono view
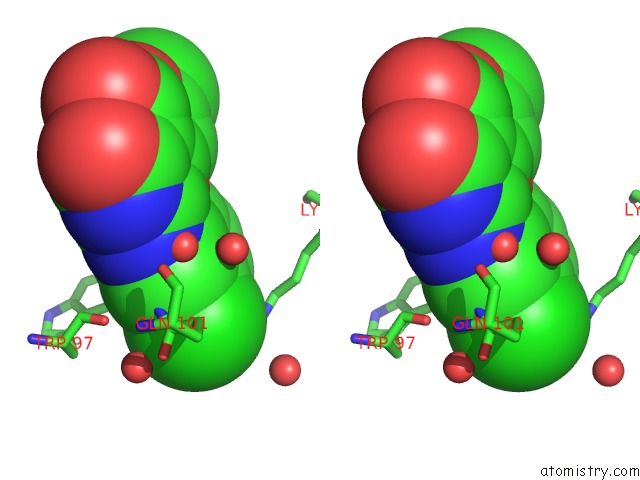
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Discovery and the Crystal Structure of NS5 in Complex with the N- Terminal Bromodomain of BRD2. within 5.0Å range:
|
Reference:
P.Deshmukh,
S.Mathur,
G.Gangadharan,
G.Krishnappa,
N.Dalavaikodihalli Nanjaiah,
B.Padmanabhan.
Novel Pyrano 1,3 Oxazine Based Ligand Inhibits the Epigenetic Reader HBRD2 in Glioblastoma. Biochem.J. V. 477 2263 2020.
ISSN: ESSN 1470-8728
PubMed: 32484211
DOI: 10.1042/BCJ20200339
Page generated: Sat Jul 12 16:04:36 2025
ISSN: ESSN 1470-8728
PubMed: 32484211
DOI: 10.1042/BCJ20200339
Last articles
Mn in 7U76Mn in 7U7E
Mn in 7U7D
Mn in 7U5N
Mn in 7U74
Mn in 7U75
Mn in 7U60
Mn in 7TXA
Mn in 7U73
Mn in 7TZU