Chlorine »
PDB 6m8q-6mjh »
6mbe »
Chlorine in PDB 6mbe: Human Mcl-1 in Complex with the Designed Peptide DM7
Protein crystallography data
The structure of Human Mcl-1 in Complex with the Designed Peptide DM7, PDB code: 6mbe
was solved by
J.M.Jenson,
A.E.Keating,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 29.94 / 2.25 |
| Space group | P 32 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 80.758, 80.758, 57.950, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 19.2 / 22.1 |
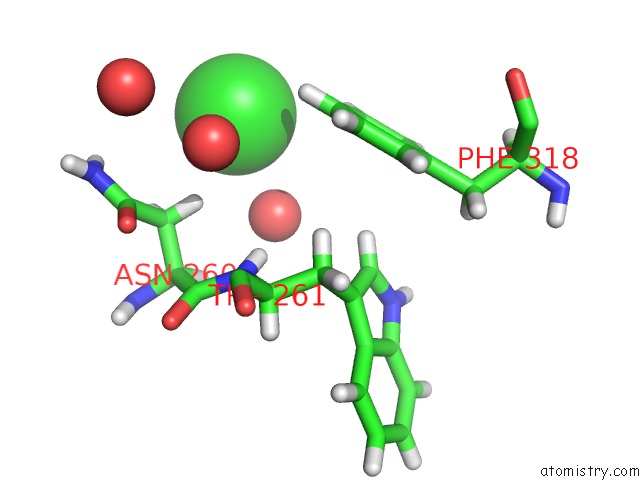
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Human Mcl-1 in Complex with the Designed Peptide DM7
(pdb code 6mbe). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total only one binding site of Chlorine was determined in the Human Mcl-1 in Complex with the Designed Peptide DM7, PDB code: 6mbe:
In total only one binding site of Chlorine was determined in the Human Mcl-1 in Complex with the Designed Peptide DM7, PDB code: 6mbe:
Chlorine binding site 1 out of 1 in 6mbe
Go back to
Chlorine binding site 1 out
of 1 in the Human Mcl-1 in Complex with the Designed Peptide DM7
Mono view
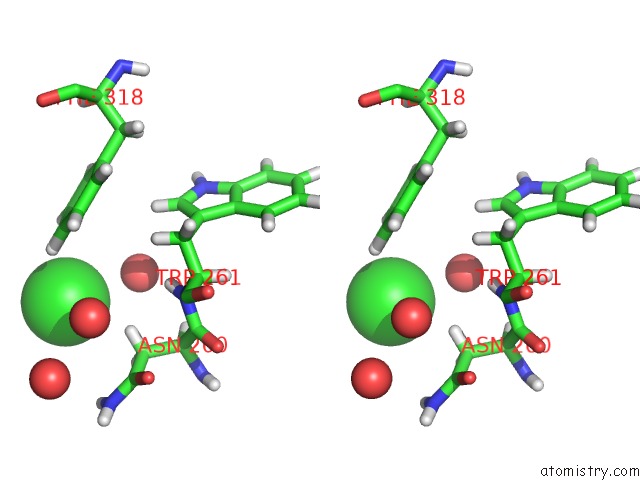
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Human Mcl-1 in Complex with the Designed Peptide DM7 within 5.0Å range:
|
Reference:
V.Frappier,
J.M.Jenson,
J.Zhou,
G.Grigoryan,
A.E.Keating.
Tertiary Structural Motif Sequence Statistics Enable Facile Prediction and Design of Peptides That Bind Anti-Apoptotic Bfl-1 and Mcl-1. Structure V. 27 606 2019.
ISSN: ISSN 1878-4186
PubMed: 30773399
DOI: 10.1016/J.STR.2019.01.008
Page generated: Sat Jul 12 16:45:22 2025
ISSN: ISSN 1878-4186
PubMed: 30773399
DOI: 10.1016/J.STR.2019.01.008
Last articles
Zn in 5A7MZn in 5ABA
Zn in 5AB2
Zn in 5AB9
Zn in 5AB0
Zn in 5AAZ
Zn in 5AAS
Zn in 5AAY
Zn in 5A87
Zn in 5AAQ