Chlorine »
PDB 6mr9-6n3x »
6mtn »
Chlorine in PDB 6mtn: Crystal Structure of Hiv-1 BG505 Sosip.664 Prefusion Env Trimer Bound to Small Molecule Hiv-1 Entry Inhibitor Compound 484 in Complex with Human Antibodies 3H109L and 35O22 at 3.0 Angstrom
Protein crystallography data
The structure of Crystal Structure of Hiv-1 BG505 Sosip.664 Prefusion Env Trimer Bound to Small Molecule Hiv-1 Entry Inhibitor Compound 484 in Complex with Human Antibodies 3H109L and 35O22 at 3.0 Angstrom, PDB code: 6mtn
was solved by
Y.-T.Lai,
P.D.Kwong,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 43.04 / 2.50 |
| Space group | P 63 |
| Cell size a, b, c (Å), α, β, γ (°) | 131.500, 131.500, 315.740, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 22.3 / 26.3 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Crystal Structure of Hiv-1 BG505 Sosip.664 Prefusion Env Trimer Bound to Small Molecule Hiv-1 Entry Inhibitor Compound 484 in Complex with Human Antibodies 3H109L and 35O22 at 3.0 Angstrom
(pdb code 6mtn). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total only one binding site of Chlorine was determined in the Crystal Structure of Hiv-1 BG505 Sosip.664 Prefusion Env Trimer Bound to Small Molecule Hiv-1 Entry Inhibitor Compound 484 in Complex with Human Antibodies 3H109L and 35O22 at 3.0 Angstrom, PDB code: 6mtn:
In total only one binding site of Chlorine was determined in the Crystal Structure of Hiv-1 BG505 Sosip.664 Prefusion Env Trimer Bound to Small Molecule Hiv-1 Entry Inhibitor Compound 484 in Complex with Human Antibodies 3H109L and 35O22 at 3.0 Angstrom, PDB code: 6mtn:
Chlorine binding site 1 out of 1 in 6mtn
Go back to
Chlorine binding site 1 out
of 1 in the Crystal Structure of Hiv-1 BG505 Sosip.664 Prefusion Env Trimer Bound to Small Molecule Hiv-1 Entry Inhibitor Compound 484 in Complex with Human Antibodies 3H109L and 35O22 at 3.0 Angstrom
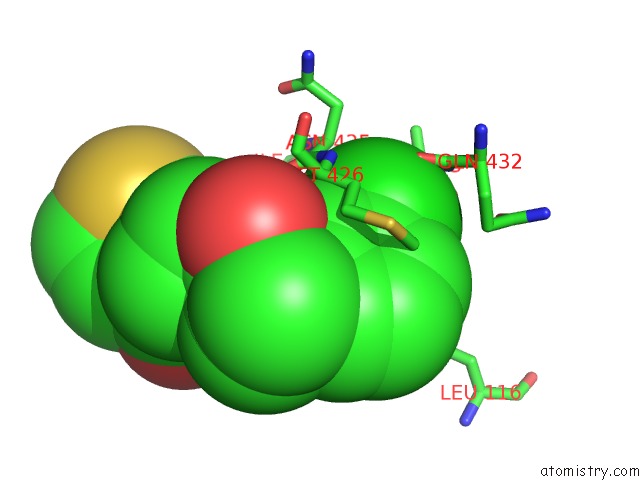
Mono view
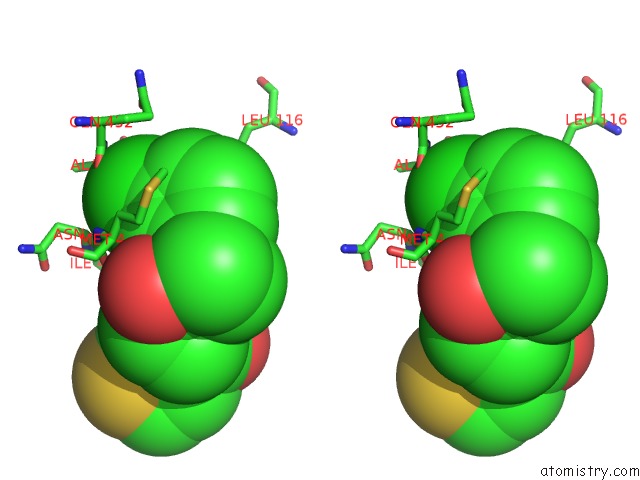
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Crystal Structure of Hiv-1 BG505 Sosip.664 Prefusion Env Trimer Bound to Small Molecule Hiv-1 Entry Inhibitor Compound 484 in Complex with Human Antibodies 3H109L and 35O22 at 3.0 Angstrom within 5.0Å range:
|
Reference:
Y.T.Lai,
T.Wang,
S.O'dell,
M.K.Louder,
A.Schon,
C.S.F.Cheung,
G.Y.Chuang,
A.Druz,
B.Lin,
K.Mckee,
D.Peng,
Y.Yang,
B.Zhang,
A.Herschhorn,
J.Sodroski,
R.T.Bailer,
N.A.Doria-Rose,
J.R.Mascola,
D.R.Langley,
P.D.Kwong.
Lattice Engineering Enables Definition of Molecular Features Allowing For Potent Small-Molecule Inhibition of Hiv-1 Entry. Nat Commun V. 10 47 2019.
ISSN: ESSN 2041-1723
PubMed: 30604750
DOI: 10.1038/S41467-018-07851-1
Page generated: Sat Jul 12 16:56:24 2025
ISSN: ESSN 2041-1723
PubMed: 30604750
DOI: 10.1038/S41467-018-07851-1
Last articles
Fe in 9JQDFe in 9KPP
Fe in 9KGP
Fe in 9K3V
Fe in 9K3U
Fe in 9KDG
Fe in 9KDF
Fe in 9K3B
Fe in 9K3Q
Fe in 9K39