Chlorine »
PDB 6n3y-6ndf »
6nad »
Chlorine in PDB 6nad: Identification and Biological Evaluation of Tertiary Alcohol-Based Inverse Agonists of Rorgt
Protein crystallography data
The structure of Identification and Biological Evaluation of Tertiary Alcohol-Based Inverse Agonists of Rorgt, PDB code: 6nad
was solved by
J.Spurlino,
C.Milligan,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 48.37 / 2.90 |
| Space group | P 61 |
| Cell size a, b, c (Å), α, β, γ (°) | 96.742, 96.742, 127.711, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 17.6 / 23.4 |
Other elements in 6nad:
The structure of Identification and Biological Evaluation of Tertiary Alcohol-Based Inverse Agonists of Rorgt also contains other interesting chemical elements:
| Fluorine | (F) | 12 atoms |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Identification and Biological Evaluation of Tertiary Alcohol-Based Inverse Agonists of Rorgt
(pdb code 6nad). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 2 binding sites of Chlorine where determined in the Identification and Biological Evaluation of Tertiary Alcohol-Based Inverse Agonists of Rorgt, PDB code: 6nad:
Jump to Chlorine binding site number: 1; 2;
In total 2 binding sites of Chlorine where determined in the Identification and Biological Evaluation of Tertiary Alcohol-Based Inverse Agonists of Rorgt, PDB code: 6nad:
Jump to Chlorine binding site number: 1; 2;
Chlorine binding site 1 out of 2 in 6nad
Go back to
Chlorine binding site 1 out
of 2 in the Identification and Biological Evaluation of Tertiary Alcohol-Based Inverse Agonists of Rorgt
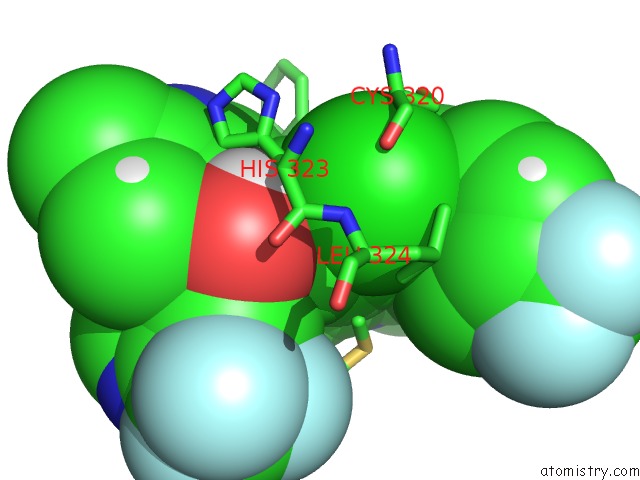
Mono view
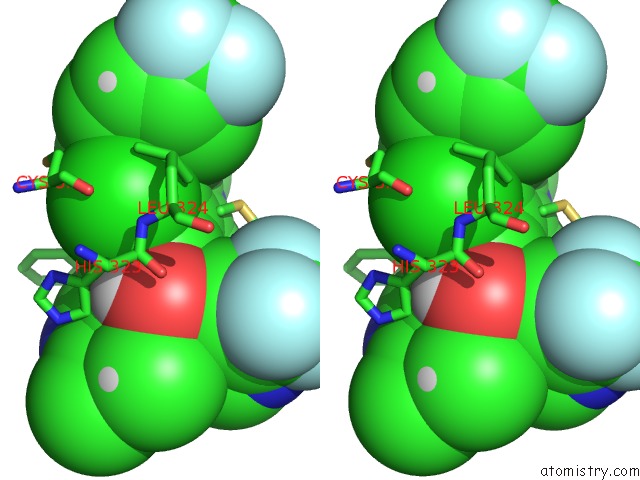
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Identification and Biological Evaluation of Tertiary Alcohol-Based Inverse Agonists of Rorgt within 5.0Å range:
|
Chlorine binding site 2 out of 2 in 6nad
Go back to
Chlorine binding site 2 out
of 2 in the Identification and Biological Evaluation of Tertiary Alcohol-Based Inverse Agonists of Rorgt
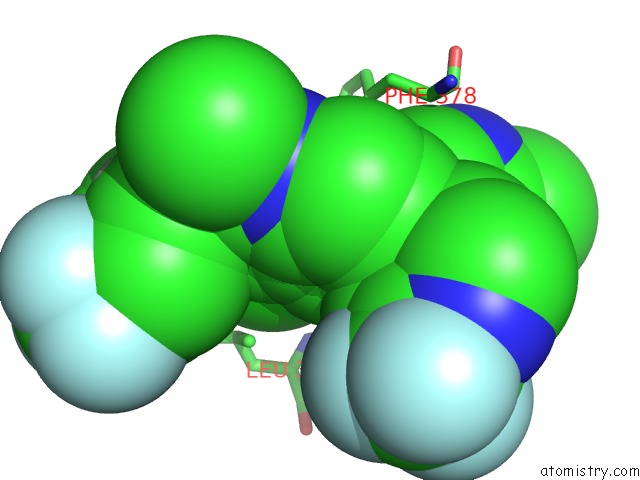
Mono view
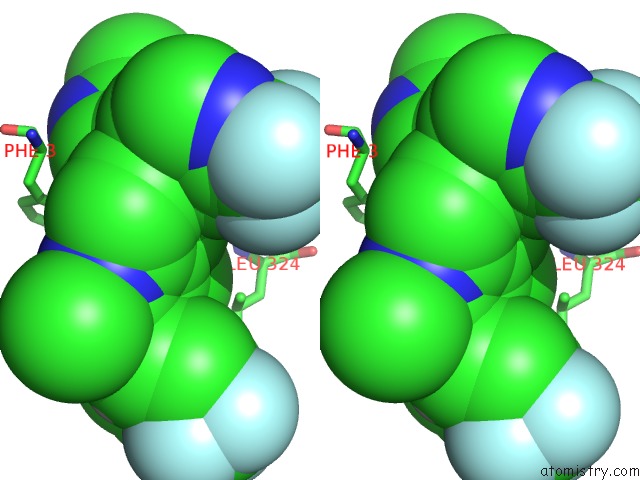
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Identification and Biological Evaluation of Tertiary Alcohol-Based Inverse Agonists of Rorgt within 5.0Å range:
|
Reference:
V.M.Tanis,
H.Venkatesan,
M.D.Cummings,
M.Albers,
J.Kent Barbay,
K.Herman,
D.A.Kummer,
C.Milligan,
M.I.Nelen,
R.Nishimura,
T.Schlueter,
B.Scott,
J.Spurlino,
R.Wolin,
C.Woods,
X.Xue,
J.P.Edwards,
A.M.Fourie,
K.Leonard.
3-Substituted Quinolines As Ror Gamma T Inverse Agonists. Bioorg.Med.Chem.Lett. V. 29 1463 2019.
ISSN: ESSN 1464-3405
PubMed: 31010722
DOI: 10.1016/J.BMCL.2019.04.021
Page generated: Sat Jul 12 17:05:03 2025
ISSN: ESSN 1464-3405
PubMed: 31010722
DOI: 10.1016/J.BMCL.2019.04.021
Last articles
K in 3GKOK in 3GVF
K in 3GJK
K in 3GE2
K in 3GAI
K in 3G71
K in 3GAJ
K in 3G6E
K in 3GAH
K in 3FWP