Chlorine »
PDB 6ouy-6p2l »
6p02 »
Chlorine in PDB 6p02: Crystal Structure of Mtb Aspartate Decarboxylase, 6-Chlorine Pyrazinoic Acid Complex
Enzymatic activity of Crystal Structure of Mtb Aspartate Decarboxylase, 6-Chlorine Pyrazinoic Acid Complex
All present enzymatic activity of Crystal Structure of Mtb Aspartate Decarboxylase, 6-Chlorine Pyrazinoic Acid Complex:
4.1.1.11;
4.1.1.11;
Protein crystallography data
The structure of Crystal Structure of Mtb Aspartate Decarboxylase, 6-Chlorine Pyrazinoic Acid Complex, PDB code: 6p02
was solved by
Q.Sun,
X.Li,
J.C.Sacchettini,
Tb Structural Genomics Consortium (Tbsgc),
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 21.89 / 2.25 |
| Space group | P 31 |
| Cell size a, b, c (Å), α, β, γ (°) | 143.679, 143.679, 59.838, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 13.4 / 16.4 |
Chlorine Binding Sites:
Pages:
>>> Page 1 <<< Page 2, Binding sites: 11 - 12;Binding sites:
The binding sites of Chlorine atom in the Crystal Structure of Mtb Aspartate Decarboxylase, 6-Chlorine Pyrazinoic Acid Complex (pdb code 6p02). This binding sites where shown within 5.0 Angstroms radius around Chlorine atom.In total 12 binding sites of Chlorine where determined in the Crystal Structure of Mtb Aspartate Decarboxylase, 6-Chlorine Pyrazinoic Acid Complex, PDB code: 6p02:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5; 6; 7; 8; 9; 10;
Chlorine binding site 1 out of 12 in 6p02
Go back to
Chlorine binding site 1 out
of 12 in the Crystal Structure of Mtb Aspartate Decarboxylase, 6-Chlorine Pyrazinoic Acid Complex
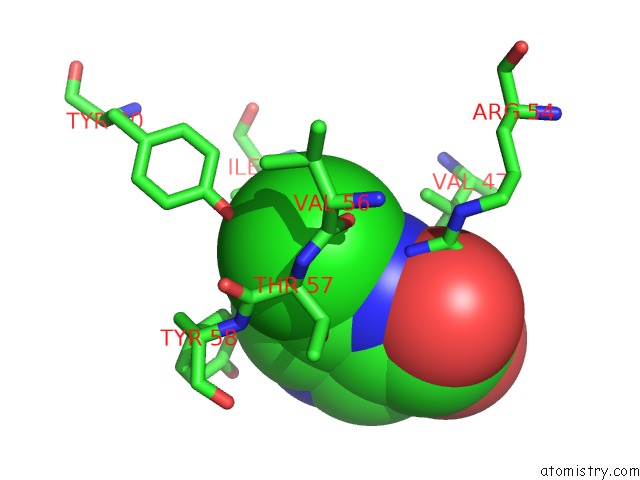
Mono view
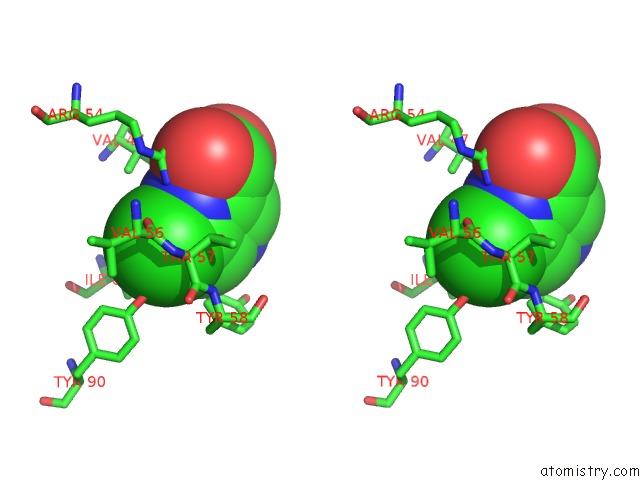
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Crystal Structure of Mtb Aspartate Decarboxylase, 6-Chlorine Pyrazinoic Acid Complex within 5.0Å range:
|
Chlorine binding site 2 out of 12 in 6p02
Go back to
Chlorine binding site 2 out
of 12 in the Crystal Structure of Mtb Aspartate Decarboxylase, 6-Chlorine Pyrazinoic Acid Complex
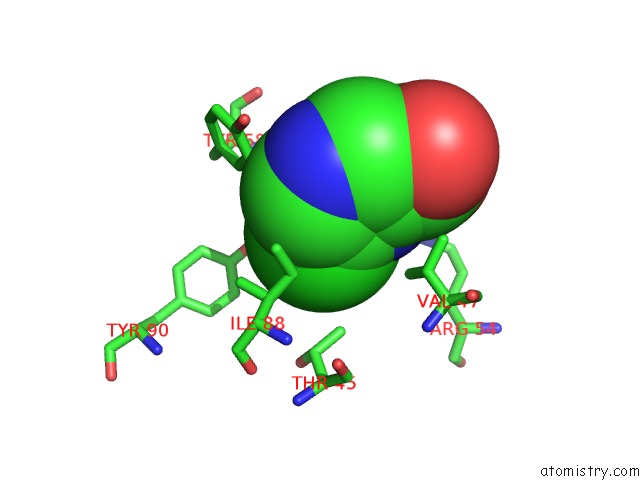
Mono view
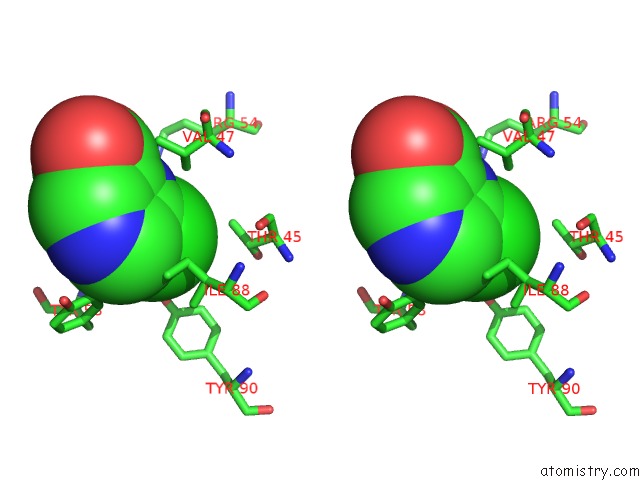
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Crystal Structure of Mtb Aspartate Decarboxylase, 6-Chlorine Pyrazinoic Acid Complex within 5.0Å range:
|
Chlorine binding site 3 out of 12 in 6p02
Go back to
Chlorine binding site 3 out
of 12 in the Crystal Structure of Mtb Aspartate Decarboxylase, 6-Chlorine Pyrazinoic Acid Complex
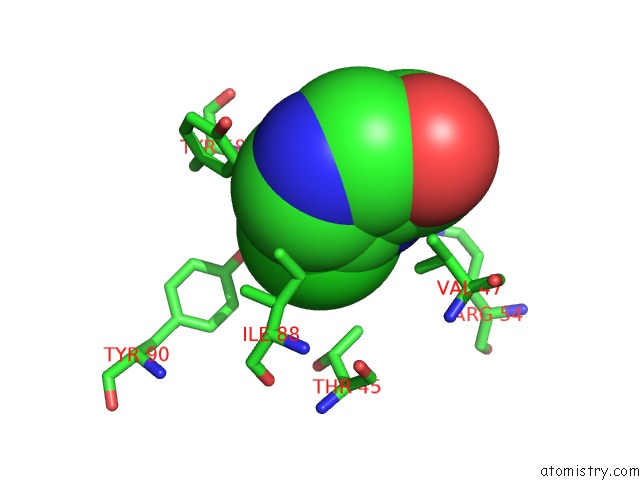
Mono view
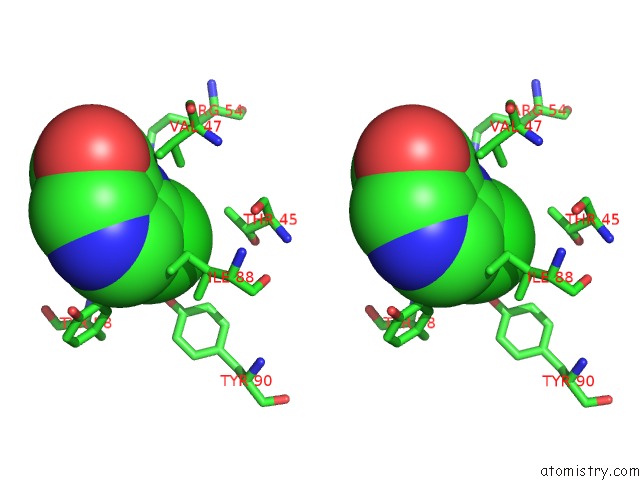
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Crystal Structure of Mtb Aspartate Decarboxylase, 6-Chlorine Pyrazinoic Acid Complex within 5.0Å range:
|
Chlorine binding site 4 out of 12 in 6p02
Go back to
Chlorine binding site 4 out
of 12 in the Crystal Structure of Mtb Aspartate Decarboxylase, 6-Chlorine Pyrazinoic Acid Complex
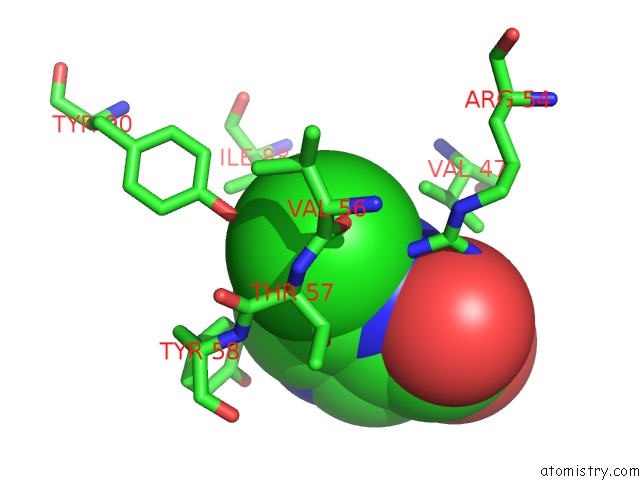
Mono view
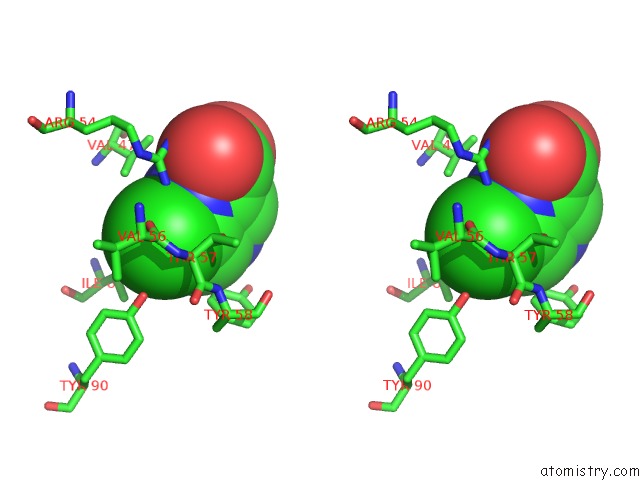
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of Crystal Structure of Mtb Aspartate Decarboxylase, 6-Chlorine Pyrazinoic Acid Complex within 5.0Å range:
|
Chlorine binding site 5 out of 12 in 6p02
Go back to
Chlorine binding site 5 out
of 12 in the Crystal Structure of Mtb Aspartate Decarboxylase, 6-Chlorine Pyrazinoic Acid Complex
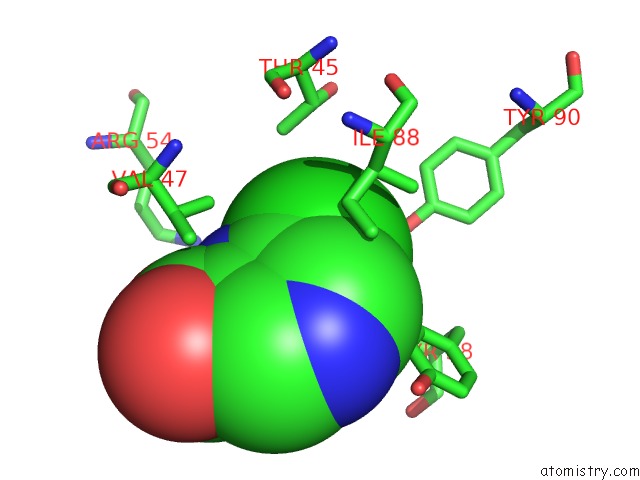
Mono view
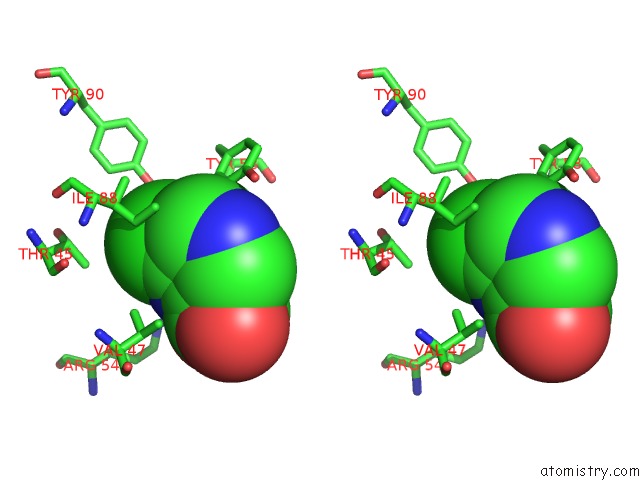
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 5 of Crystal Structure of Mtb Aspartate Decarboxylase, 6-Chlorine Pyrazinoic Acid Complex within 5.0Å range:
|
Chlorine binding site 6 out of 12 in 6p02
Go back to
Chlorine binding site 6 out
of 12 in the Crystal Structure of Mtb Aspartate Decarboxylase, 6-Chlorine Pyrazinoic Acid Complex
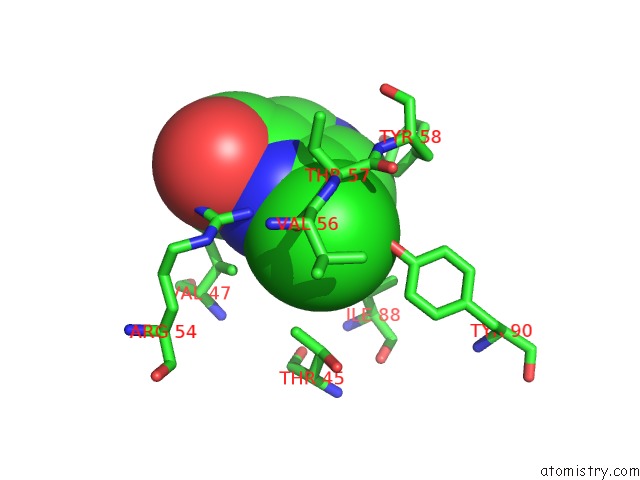
Mono view
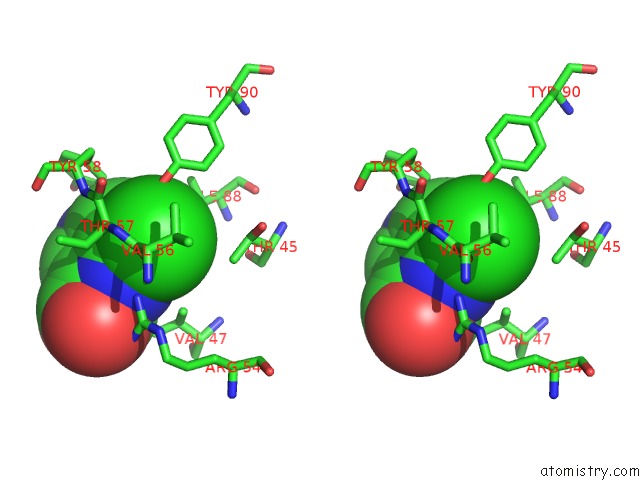
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 6 of Crystal Structure of Mtb Aspartate Decarboxylase, 6-Chlorine Pyrazinoic Acid Complex within 5.0Å range:
|
Chlorine binding site 7 out of 12 in 6p02
Go back to
Chlorine binding site 7 out
of 12 in the Crystal Structure of Mtb Aspartate Decarboxylase, 6-Chlorine Pyrazinoic Acid Complex
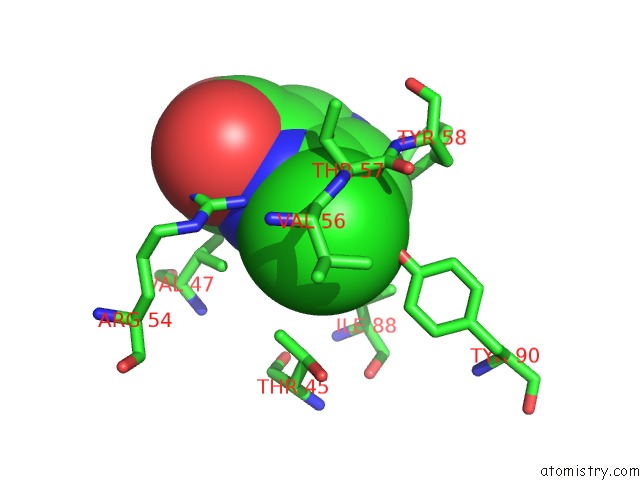
Mono view
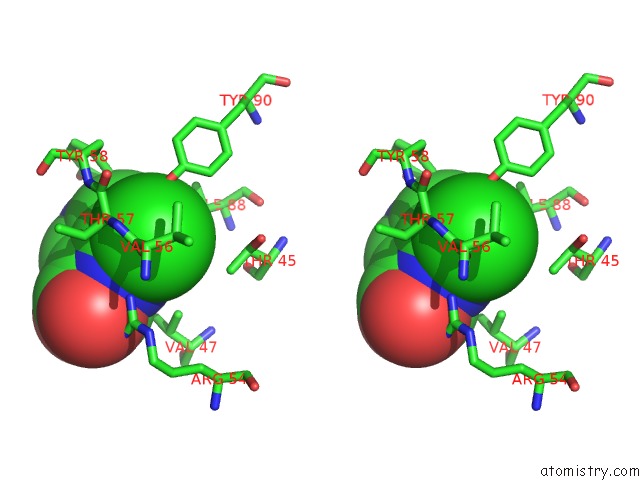
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 7 of Crystal Structure of Mtb Aspartate Decarboxylase, 6-Chlorine Pyrazinoic Acid Complex within 5.0Å range:
|
Chlorine binding site 8 out of 12 in 6p02
Go back to
Chlorine binding site 8 out
of 12 in the Crystal Structure of Mtb Aspartate Decarboxylase, 6-Chlorine Pyrazinoic Acid Complex
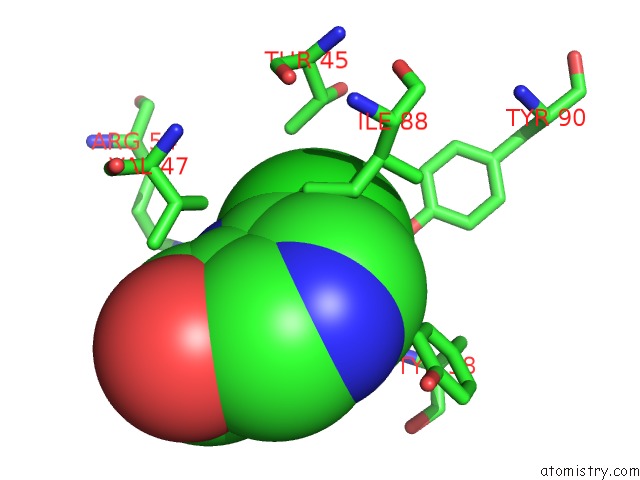
Mono view
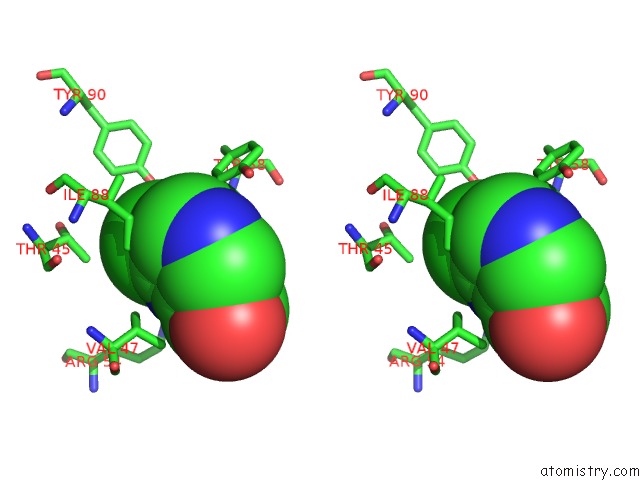
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 8 of Crystal Structure of Mtb Aspartate Decarboxylase, 6-Chlorine Pyrazinoic Acid Complex within 5.0Å range:
|
Chlorine binding site 9 out of 12 in 6p02
Go back to
Chlorine binding site 9 out
of 12 in the Crystal Structure of Mtb Aspartate Decarboxylase, 6-Chlorine Pyrazinoic Acid Complex
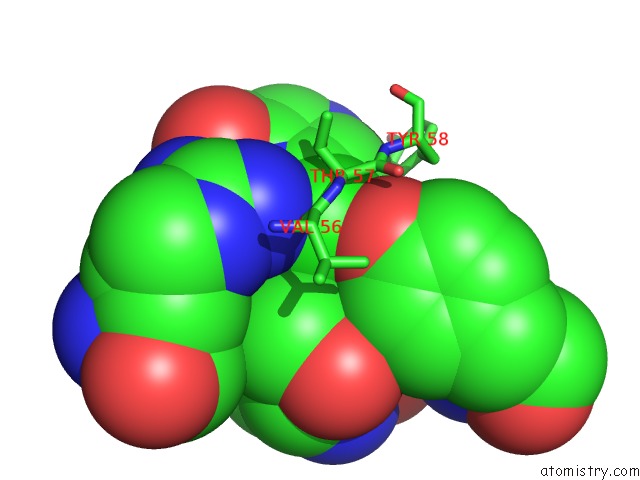
Mono view
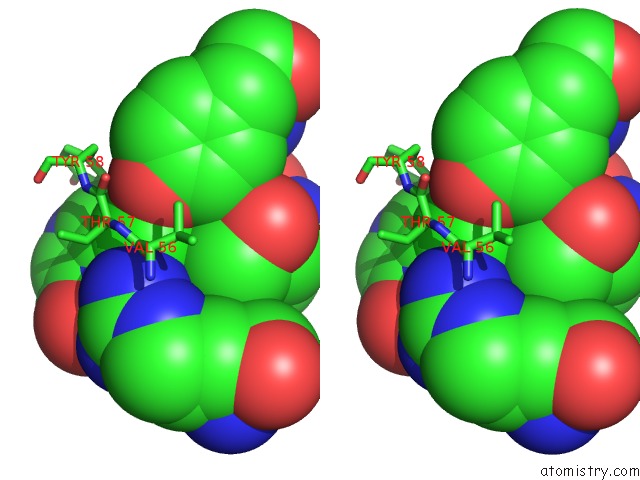
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 9 of Crystal Structure of Mtb Aspartate Decarboxylase, 6-Chlorine Pyrazinoic Acid Complex within 5.0Å range:
|
Chlorine binding site 10 out of 12 in 6p02
Go back to
Chlorine binding site 10 out
of 12 in the Crystal Structure of Mtb Aspartate Decarboxylase, 6-Chlorine Pyrazinoic Acid Complex
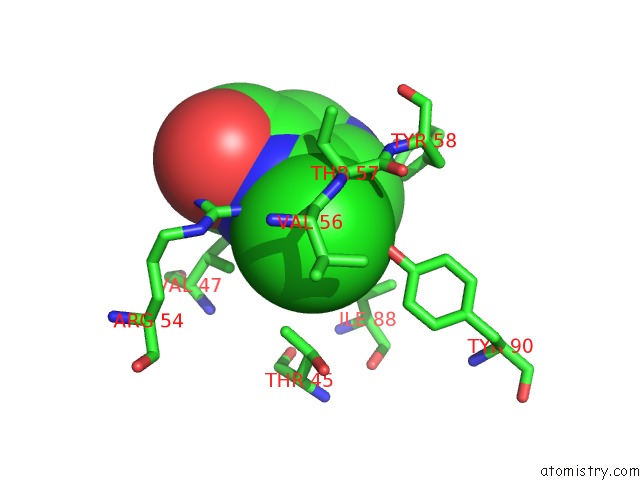
Mono view
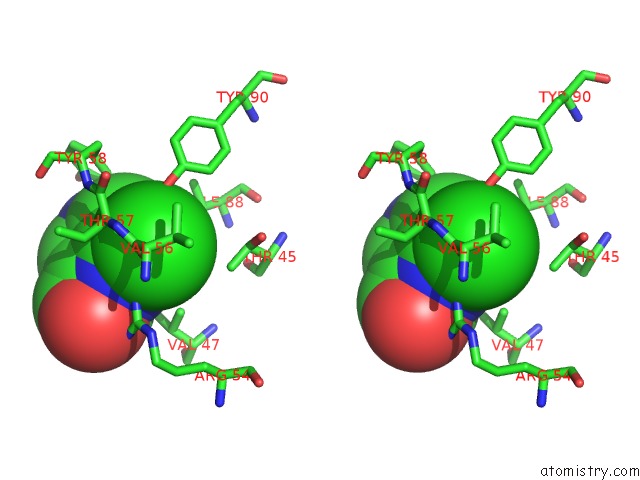
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 10 of Crystal Structure of Mtb Aspartate Decarboxylase, 6-Chlorine Pyrazinoic Acid Complex within 5.0Å range:
|
Reference:
Q.Sun,
X.Li,
L.M.Perez,
W.Shi,
Y.Zhang,
J.C.Sacchettini.
The Molecular Basis of Pyrazinamide Activity on Mycobacterium Tuberculosis Pand. Nat Commun V. 11 339 2020.
ISSN: ESSN 2041-1723
PubMed: 31953389
DOI: 10.1038/S41467-019-14238-3
Page generated: Sat Jul 12 18:03:23 2025
ISSN: ESSN 2041-1723
PubMed: 31953389
DOI: 10.1038/S41467-019-14238-3
Last articles
Mg in 7DUJMg in 7DUI
Mg in 7DUH
Mg in 7DUG
Mg in 7DR0
Mg in 7DR1
Mg in 7DU2
Mg in 7DSP
Mg in 7DSJ
Mg in 7DSI