Chlorine »
PDB 6prr-6q0q »
6pry »
Chlorine in PDB 6pry: X-Ray Crystal Structure of the Blue-Light Absorbing State of Pixj From Thermosynechococcus Elongatus By Serial Femtosecond Crystallographic Analysis
Protein crystallography data
The structure of X-Ray Crystal Structure of the Blue-Light Absorbing State of Pixj From Thermosynechococcus Elongatus By Serial Femtosecond Crystallographic Analysis, PDB code: 6pry
was solved by
E.S.Burgie,
J.A.Clinger,
M.D.Miller,
G.N.Phillips Jr.,
R.D.Vierstra,
A.M.Orville,
J.F.Kern,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 24.09 / 1.55 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 44.788, 61.417, 116.484, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 15.7 / 18.5 |
Other elements in 6pry:
The structure of X-Ray Crystal Structure of the Blue-Light Absorbing State of Pixj From Thermosynechococcus Elongatus By Serial Femtosecond Crystallographic Analysis also contains other interesting chemical elements:
| Magnesium | (Mg) | 4 atoms |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the X-Ray Crystal Structure of the Blue-Light Absorbing State of Pixj From Thermosynechococcus Elongatus By Serial Femtosecond Crystallographic Analysis
(pdb code 6pry). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 3 binding sites of Chlorine where determined in the X-Ray Crystal Structure of the Blue-Light Absorbing State of Pixj From Thermosynechococcus Elongatus By Serial Femtosecond Crystallographic Analysis, PDB code: 6pry:
Jump to Chlorine binding site number: 1; 2; 3;
In total 3 binding sites of Chlorine where determined in the X-Ray Crystal Structure of the Blue-Light Absorbing State of Pixj From Thermosynechococcus Elongatus By Serial Femtosecond Crystallographic Analysis, PDB code: 6pry:
Jump to Chlorine binding site number: 1; 2; 3;
Chlorine binding site 1 out of 3 in 6pry
Go back to
Chlorine binding site 1 out
of 3 in the X-Ray Crystal Structure of the Blue-Light Absorbing State of Pixj From Thermosynechococcus Elongatus By Serial Femtosecond Crystallographic Analysis
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of X-Ray Crystal Structure of the Blue-Light Absorbing State of Pixj From Thermosynechococcus Elongatus By Serial Femtosecond Crystallographic Analysis within 5.0Å range:
|
Chlorine binding site 2 out of 3 in 6pry
Go back to
Chlorine binding site 2 out
of 3 in the X-Ray Crystal Structure of the Blue-Light Absorbing State of Pixj From Thermosynechococcus Elongatus By Serial Femtosecond Crystallographic Analysis
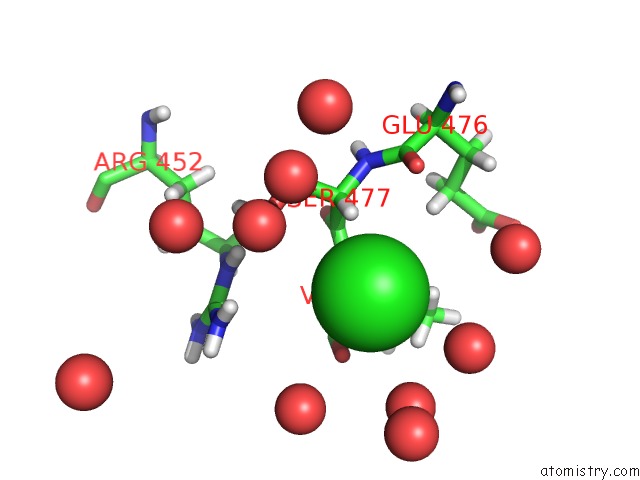
Mono view
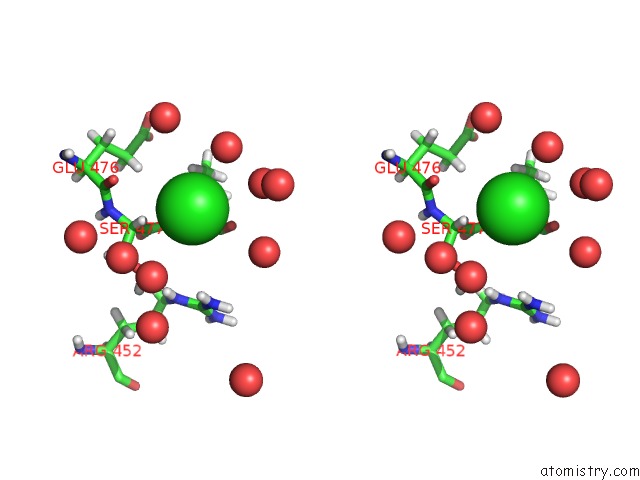
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of X-Ray Crystal Structure of the Blue-Light Absorbing State of Pixj From Thermosynechococcus Elongatus By Serial Femtosecond Crystallographic Analysis within 5.0Å range:
|
Chlorine binding site 3 out of 3 in 6pry
Go back to
Chlorine binding site 3 out
of 3 in the X-Ray Crystal Structure of the Blue-Light Absorbing State of Pixj From Thermosynechococcus Elongatus By Serial Femtosecond Crystallographic Analysis

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of X-Ray Crystal Structure of the Blue-Light Absorbing State of Pixj From Thermosynechococcus Elongatus By Serial Femtosecond Crystallographic Analysis within 5.0Å range:
|
Reference:
E.S.Burgie,
J.A.Clinger,
M.D.Miller,
A.S.Brewster,
P.Aller,
A.Butryn,
F.D.Fuller,
S.Gul,
I.D.Young,
C.C.Pham,
I.S.Kim,
A.Bhowmick,
L.J.O'riordan,
K.D.Sutherlin,
J.V.Heinemann,
A.Batyuk,
R.Alonso-Mori,
M.S.Hunter,
J.E.Koglin,
J.Yano,
V.K.Yachandra,
N.K.Sauter,
A.E.Cohen,
J.Kern,
A.M.Orville,
G.N.Phillips Jr.,
R.D.Vierstra.
Photoreversible Interconversion of A Phytochrome Photosensory Module in the Crystalline State. Proc.Natl.Acad.Sci.Usa 2019.
ISSN: ESSN 1091-6490
PubMed: 31852825
DOI: 10.1073/PNAS.1912041116
Page generated: Sat Jul 12 18:34:52 2025
ISSN: ESSN 1091-6490
PubMed: 31852825
DOI: 10.1073/PNAS.1912041116
Last articles
I in 5DLTI in 5CYM
I in 5D2R
I in 5DIL
I in 5DLI
I in 5DL9
I in 5DJC
I in 5D69
I in 5DAM
I in 5CW1