Chlorine »
PDB 6sj7-6sr4 »
6sr1 »
Chlorine in PDB 6sr1: X-Ray Pump X-Ray Probe on Lysozyme.Gd Nanocrystals: 35 Fs Time Delay
Enzymatic activity of X-Ray Pump X-Ray Probe on Lysozyme.Gd Nanocrystals: 35 Fs Time Delay
All present enzymatic activity of X-Ray Pump X-Ray Probe on Lysozyme.Gd Nanocrystals: 35 Fs Time Delay:
3.2.1.17;
3.2.1.17;
Protein crystallography data
The structure of X-Ray Pump X-Ray Probe on Lysozyme.Gd Nanocrystals: 35 Fs Time Delay, PDB code: 6sr1
was solved by
M.Kloos,
A.Gorel,
K.Nass,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 22.81 / 2.30 |
| Space group | P 43 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 79.000, 79.000, 39.500, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 17.9 / 24.5 |
Other elements in 6sr1:
The structure of X-Ray Pump X-Ray Probe on Lysozyme.Gd Nanocrystals: 35 Fs Time Delay also contains other interesting chemical elements:
| Gadolinium | (Gd) | 2 atoms |
| Sodium | (Na) | 1 atom |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the X-Ray Pump X-Ray Probe on Lysozyme.Gd Nanocrystals: 35 Fs Time Delay
(pdb code 6sr1). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 3 binding sites of Chlorine where determined in the X-Ray Pump X-Ray Probe on Lysozyme.Gd Nanocrystals: 35 Fs Time Delay, PDB code: 6sr1:
Jump to Chlorine binding site number: 1; 2; 3;
In total 3 binding sites of Chlorine where determined in the X-Ray Pump X-Ray Probe on Lysozyme.Gd Nanocrystals: 35 Fs Time Delay, PDB code: 6sr1:
Jump to Chlorine binding site number: 1; 2; 3;
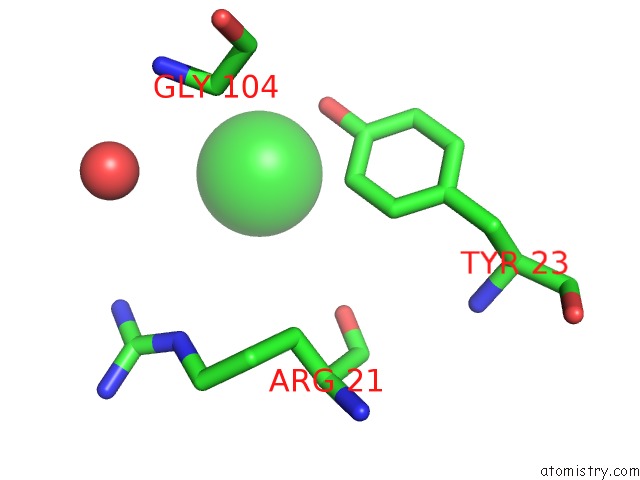
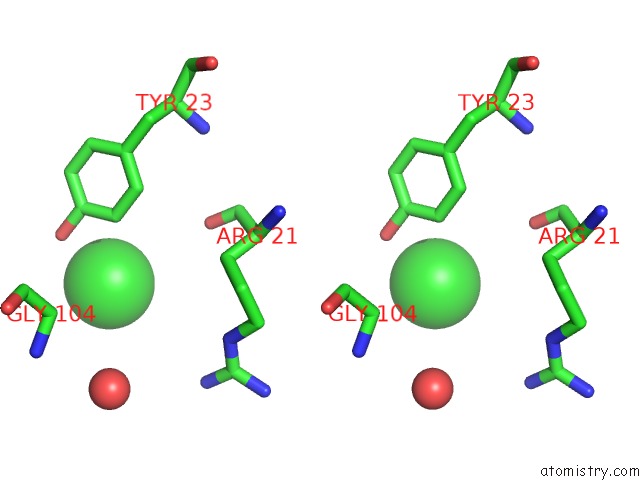
Chlorine binding site 1 out of 3 in 6sr1
Go back to
Chlorine binding site 1 out
of 3 in the X-Ray Pump X-Ray Probe on Lysozyme.Gd Nanocrystals: 35 Fs Time Delay
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of X-Ray Pump X-Ray Probe on Lysozyme.Gd Nanocrystals: 35 Fs Time Delay within 5.0Å range:
|
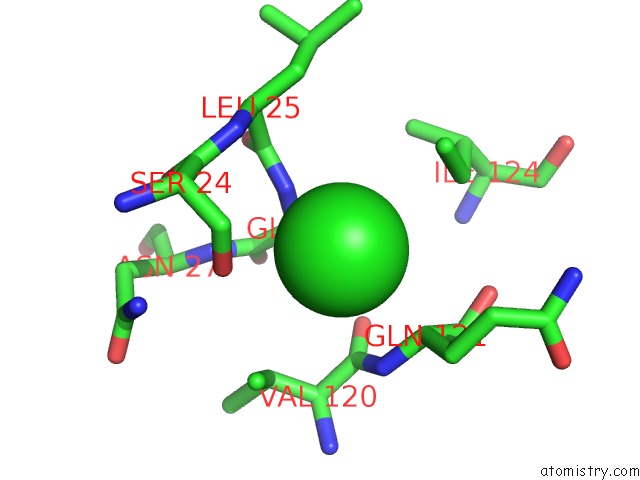
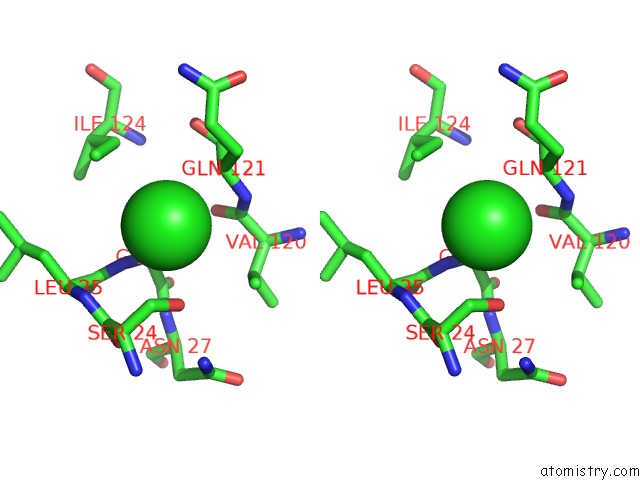
Chlorine binding site 2 out of 3 in 6sr1
Go back to
Chlorine binding site 2 out
of 3 in the X-Ray Pump X-Ray Probe on Lysozyme.Gd Nanocrystals: 35 Fs Time Delay
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of X-Ray Pump X-Ray Probe on Lysozyme.Gd Nanocrystals: 35 Fs Time Delay within 5.0Å range:
|
Chlorine binding site 3 out of 3 in 6sr1
Go back to
Chlorine binding site 3 out
of 3 in the X-Ray Pump X-Ray Probe on Lysozyme.Gd Nanocrystals: 35 Fs Time Delay
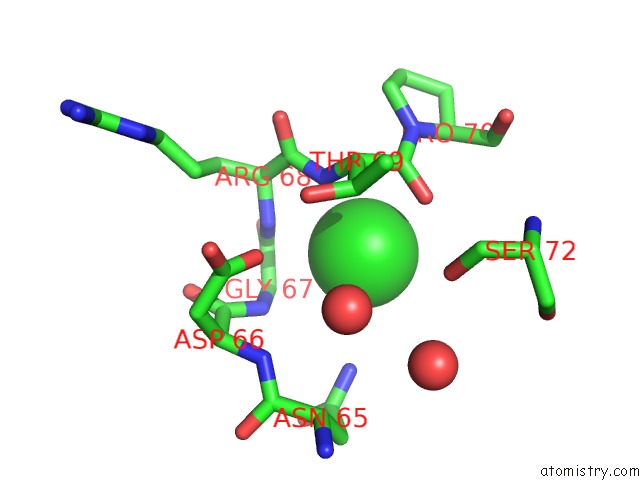
Mono view
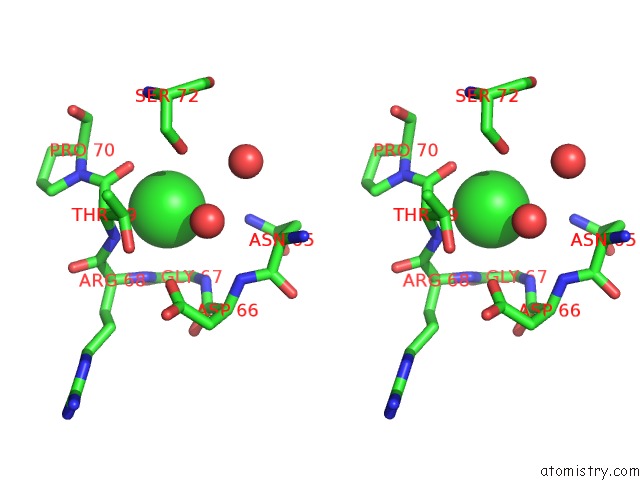
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of X-Ray Pump X-Ray Probe on Lysozyme.Gd Nanocrystals: 35 Fs Time Delay within 5.0Å range:
|
Reference:
K.Nass,
A.Gorel,
M.M.Abdullah,
A.V Martin,
M.Kloos,
A.Marinelli,
A.Aquila,
T.R.M.Barends,
F.J.Decker,
R.Bruce Doak,
L.Foucar,
E.Hartmann,
M.Hilpert,
M.S.Hunter,
Z.Jurek,
J.E.Koglin,
A.Kozlov,
A.A.Lutman,
G.N.Kovacs,
C.M.Roome,
R.L.Shoeman,
R.Santra,
H.M.Quiney,
B.Ziaja,
S.Boutet,
I.Schlichting.
Structural Dynamics in Proteins Induced By and Probed with X-Ray Free-Electron Laser Pulses. Nat Commun V. 11 1814 2020.
ISSN: ESSN 2041-1723
PubMed: 32286284
DOI: 10.1038/S41467-020-15610-4
Page generated: Sat Jul 12 19:54:52 2025
ISSN: ESSN 2041-1723
PubMed: 32286284
DOI: 10.1038/S41467-020-15610-4
Last articles
Mn in 8SUTMn in 8SUU
Mn in 8SP5
Mn in 8SND
Mn in 8SNE
Mn in 8SNC
Mn in 8SLM
Mn in 8SLN
Mn in 8SKT
Mn in 8QAV