Chlorine »
PDB 6vlm-6vx6 »
6vpy »
Chlorine in PDB 6vpy: I33M (I3.2 Mutant From CH103 Lineage)
Protein crystallography data
The structure of I33M (I3.2 Mutant From CH103 Lineage), PDB code: 6vpy
was solved by
D.Fera,
J.Zhou,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 47.65 / 2.36 |
| Space group | P 65 |
| Cell size a, b, c (Å), α, β, γ (°) | 131.605, 131.605, 104.908, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 18.5 / 22.3 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the I33M (I3.2 Mutant From CH103 Lineage)
(pdb code 6vpy). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total only one binding site of Chlorine was determined in the I33M (I3.2 Mutant From CH103 Lineage), PDB code: 6vpy:
In total only one binding site of Chlorine was determined in the I33M (I3.2 Mutant From CH103 Lineage), PDB code: 6vpy:
Chlorine binding site 1 out of 1 in 6vpy
Go back to
Chlorine binding site 1 out
of 1 in the I33M (I3.2 Mutant From CH103 Lineage)
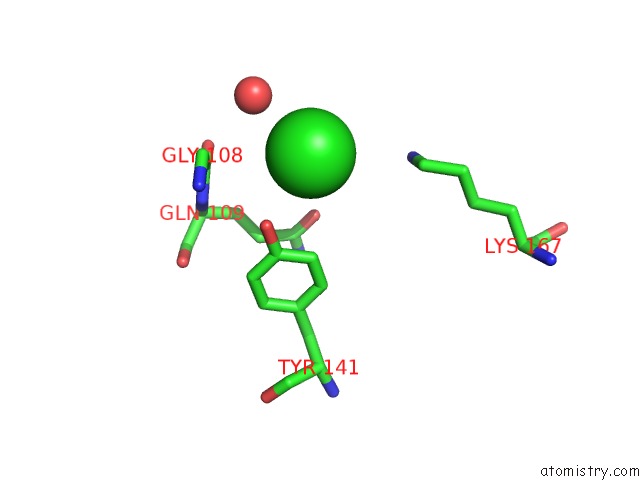
Mono view
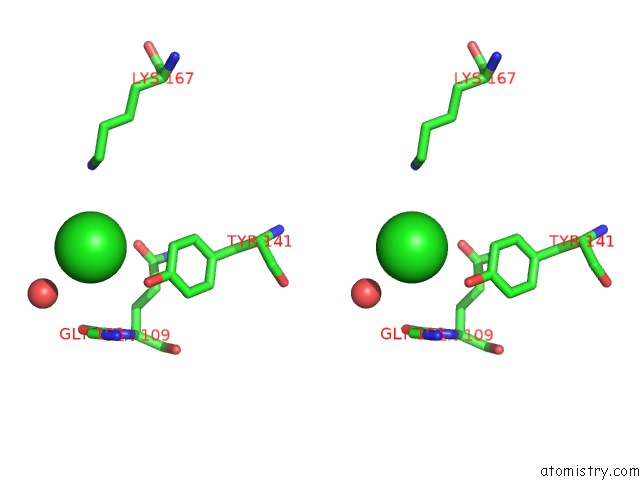
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of I33M (I3.2 Mutant From CH103 Lineage) within 5.0Å range:
|
Reference:
J.O.Zhou,
H.A.Zaidi,
T.Ton,
D.Fera.
The Effects of Framework Mutations at the Variable Domain Interface on Antibody Affinity Maturation in An Hiv-1 Broadly Neutralizing Antibody Lineage. Front Immunol V. 11 1529 2020.
ISSN: ESSN 1664-3224
PubMed: 32765530
DOI: 10.3389/FIMMU.2020.01529
Page generated: Sat Jul 12 21:02:39 2025
ISSN: ESSN 1664-3224
PubMed: 32765530
DOI: 10.3389/FIMMU.2020.01529
Last articles
Mg in 4DR2Mg in 4DR3
Mg in 4DR1
Mg in 4DPG
Mg in 4DQP
Mg in 4DQQ
Mg in 4DPM
Mg in 4DPV
Mg in 4DQI
Mg in 4DOB