Chlorine »
PDB 6y8a-6yga »
6y9j »
Chlorine in PDB 6y9j: Crystal Structure of Subtype-Switched Epithelial Adhesin 1 to 9 A Domain (EPA1-CBL2EPA9) From Candida Glabrata in Complex with Beta- Lactose
Protein crystallography data
The structure of Crystal Structure of Subtype-Switched Epithelial Adhesin 1 to 9 A Domain (EPA1-CBL2EPA9) From Candida Glabrata in Complex with Beta- Lactose, PDB code: 6y9j
was solved by
D.Hoffmann,
R.Diderrich,
M.Kock,
S.Friederichs,
V.Reithofer,
L.-O.Essen,
H.-U.Moesch,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 34.55 / 1.10 |
| Space group | C 2 2 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 74.220, 104.140, 69.090, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 14.6 / 16.7 |
Other elements in 6y9j:
The structure of Crystal Structure of Subtype-Switched Epithelial Adhesin 1 to 9 A Domain (EPA1-CBL2EPA9) From Candida Glabrata in Complex with Beta- Lactose also contains other interesting chemical elements:
| Calcium | (Ca) | 1 atom |
| Sodium | (Na) | 1 atom |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Crystal Structure of Subtype-Switched Epithelial Adhesin 1 to 9 A Domain (EPA1-CBL2EPA9) From Candida Glabrata in Complex with Beta- Lactose
(pdb code 6y9j). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 2 binding sites of Chlorine where determined in the Crystal Structure of Subtype-Switched Epithelial Adhesin 1 to 9 A Domain (EPA1-CBL2EPA9) From Candida Glabrata in Complex with Beta- Lactose, PDB code: 6y9j:
Jump to Chlorine binding site number: 1; 2;
In total 2 binding sites of Chlorine where determined in the Crystal Structure of Subtype-Switched Epithelial Adhesin 1 to 9 A Domain (EPA1-CBL2EPA9) From Candida Glabrata in Complex with Beta- Lactose, PDB code: 6y9j:
Jump to Chlorine binding site number: 1; 2;
Chlorine binding site 1 out of 2 in 6y9j
Go back to
Chlorine binding site 1 out
of 2 in the Crystal Structure of Subtype-Switched Epithelial Adhesin 1 to 9 A Domain (EPA1-CBL2EPA9) From Candida Glabrata in Complex with Beta- Lactose

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Crystal Structure of Subtype-Switched Epithelial Adhesin 1 to 9 A Domain (EPA1-CBL2EPA9) From Candida Glabrata in Complex with Beta- Lactose within 5.0Å range:
|
Chlorine binding site 2 out of 2 in 6y9j
Go back to
Chlorine binding site 2 out
of 2 in the Crystal Structure of Subtype-Switched Epithelial Adhesin 1 to 9 A Domain (EPA1-CBL2EPA9) From Candida Glabrata in Complex with Beta- Lactose
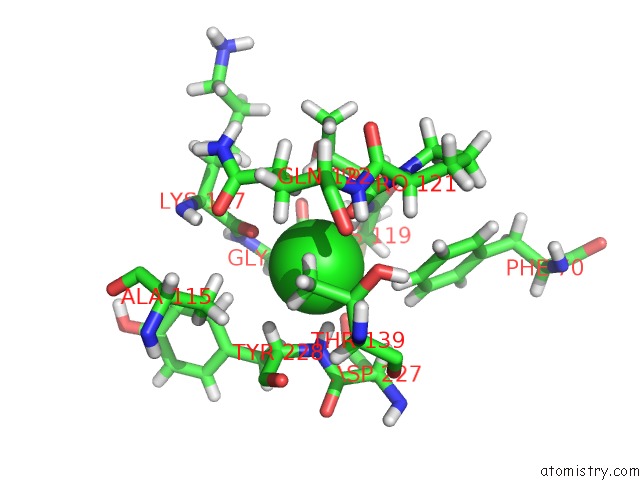
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Crystal Structure of Subtype-Switched Epithelial Adhesin 1 to 9 A Domain (EPA1-CBL2EPA9) From Candida Glabrata in Complex with Beta- Lactose within 5.0Å range:
|
Reference:
D.Hoffmann,
R.Diderrich,
V.Reithofer,
S.Friederichs,
M.Kock,
L.O.Essen,
H.U.Mosch.
Functional Reprogramming Ofcandida Glabrataepithelial Adhesins: the Role of Conserved and Variable Structural Motifs in Ligand Binding. J.Biol.Chem. V. 295 12512 2020.
ISSN: ESSN 1083-351X
PubMed: 32669365
DOI: 10.1074/JBC.RA120.013968
Page generated: Sat Jul 12 21:58:25 2025
ISSN: ESSN 1083-351X
PubMed: 32669365
DOI: 10.1074/JBC.RA120.013968
Last articles
I in 4OHMI in 4OHK
I in 4OEK
I in 4OEW
I in 4OC5
I in 4OC4
I in 4OC3
I in 4OC1
I in 4OC2
I in 4NHB