Chlorine »
PDB 7gnl-7gug »
7gs6 »
Chlorine in PDB 7gs6: Crystal Structure of Sars-Cov-2 Main Protease in Complex with Cpd-29
Enzymatic activity of Crystal Structure of Sars-Cov-2 Main Protease in Complex with Cpd-29
All present enzymatic activity of Crystal Structure of Sars-Cov-2 Main Protease in Complex with Cpd-29:
3.4.22.69;
3.4.22.69;
Protein crystallography data
The structure of Crystal Structure of Sars-Cov-2 Main Protease in Complex with Cpd-29, PDB code: 7gs6
was solved by
C.-Y.Huang,
A.Metz,
M.Sharpe,
A.Sweeney,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 56.16 / 1.62 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 67.99, 99.62, 104.44, 90, 90, 90 |
| R / Rfree (%) | 19 / 22.8 |
Other elements in 7gs6:
The structure of Crystal Structure of Sars-Cov-2 Main Protease in Complex with Cpd-29 also contains other interesting chemical elements:
| Magnesium | (Mg) | 1 atom |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Crystal Structure of Sars-Cov-2 Main Protease in Complex with Cpd-29
(pdb code 7gs6). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 6 binding sites of Chlorine where determined in the Crystal Structure of Sars-Cov-2 Main Protease in Complex with Cpd-29, PDB code: 7gs6:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5; 6;
In total 6 binding sites of Chlorine where determined in the Crystal Structure of Sars-Cov-2 Main Protease in Complex with Cpd-29, PDB code: 7gs6:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5; 6;
Chlorine binding site 1 out of 6 in 7gs6
Go back to
Chlorine binding site 1 out
of 6 in the Crystal Structure of Sars-Cov-2 Main Protease in Complex with Cpd-29
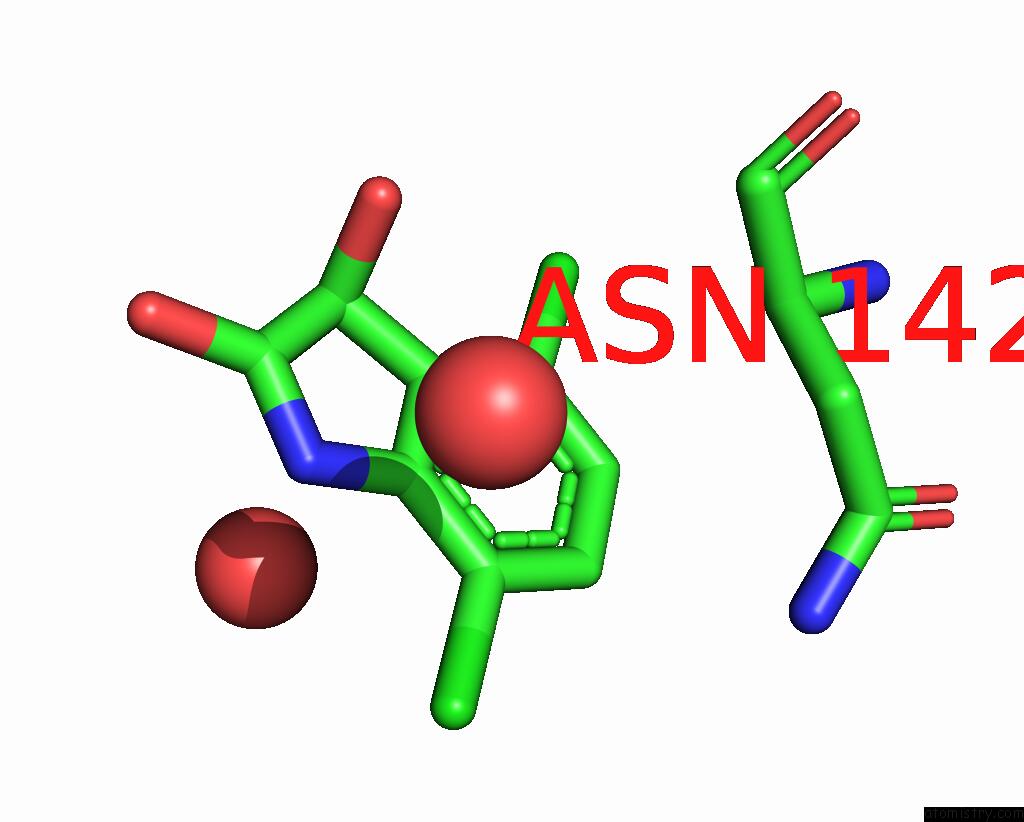
Mono view
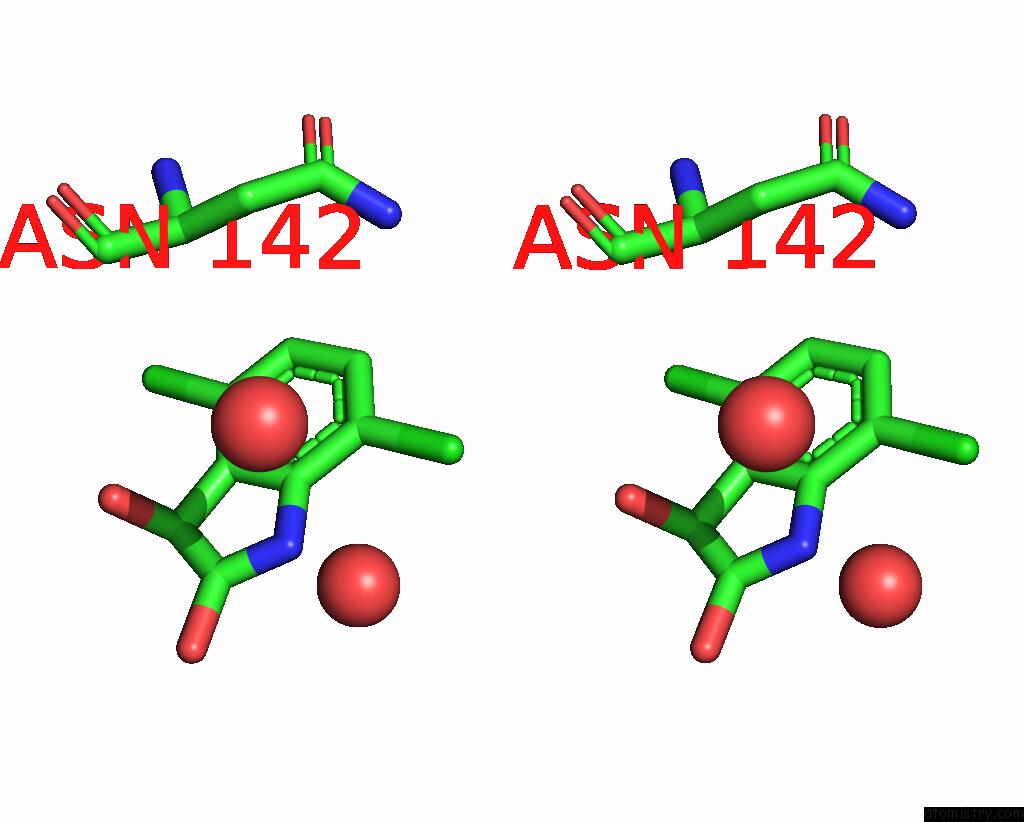
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Crystal Structure of Sars-Cov-2 Main Protease in Complex with Cpd-29 within 5.0Å range:
|
Chlorine binding site 2 out of 6 in 7gs6
Go back to
Chlorine binding site 2 out
of 6 in the Crystal Structure of Sars-Cov-2 Main Protease in Complex with Cpd-29
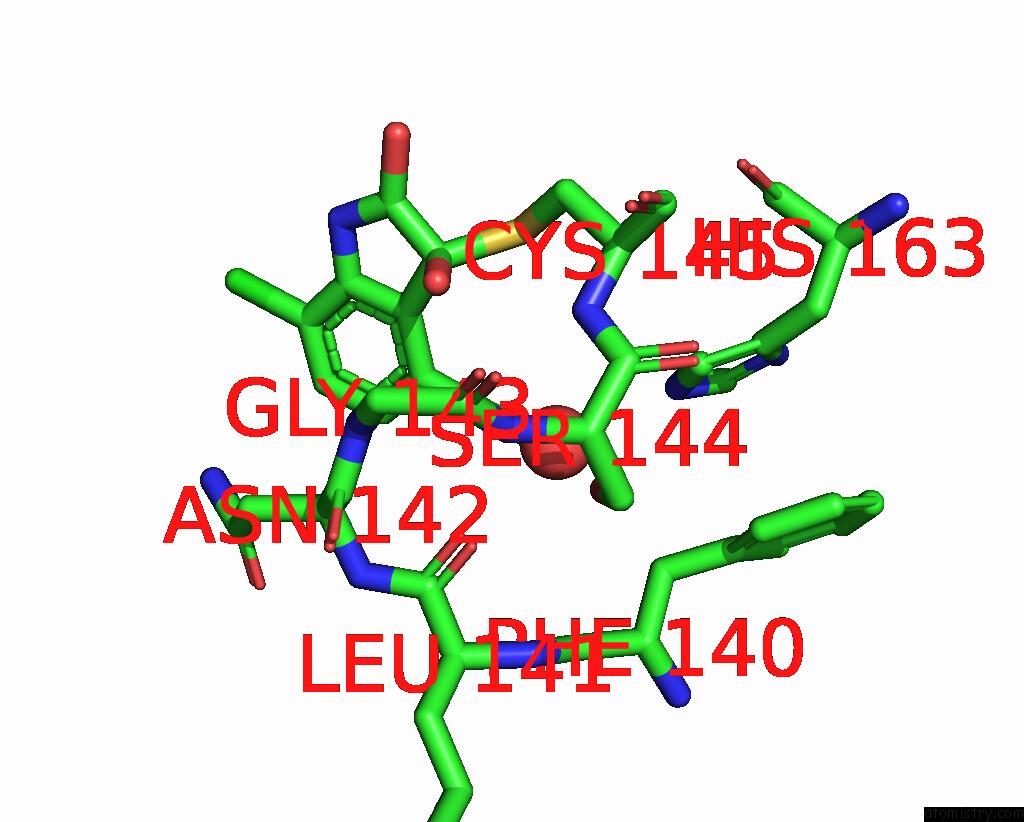
Mono view
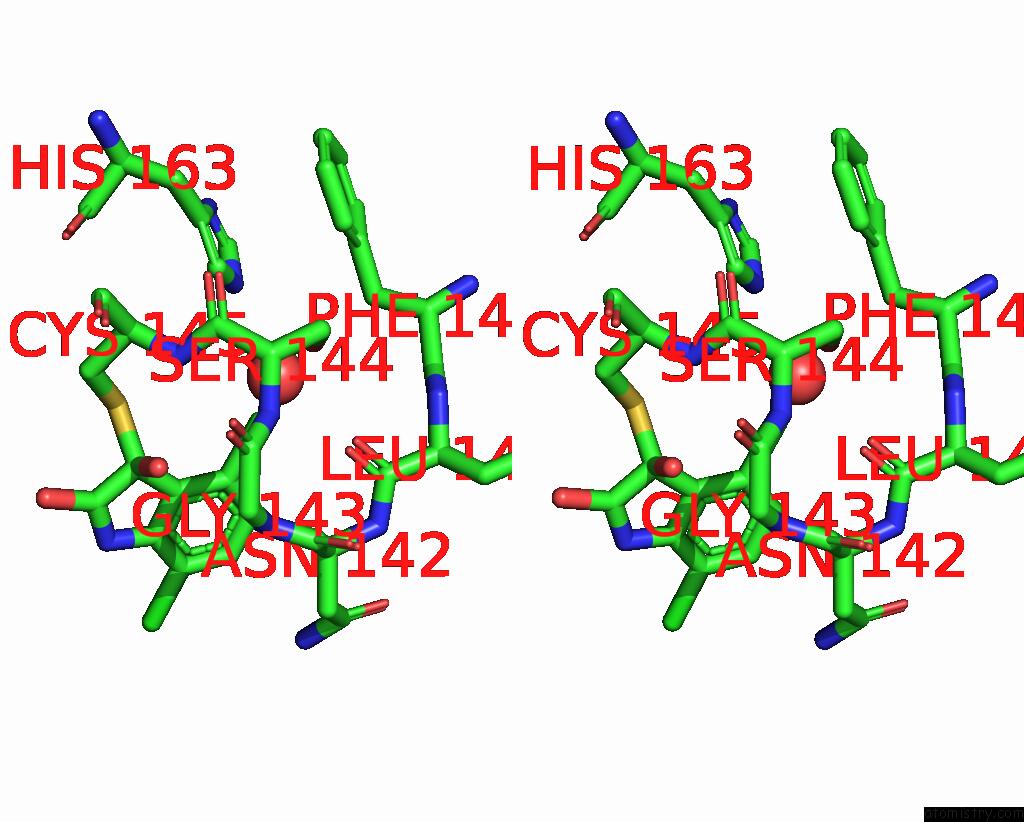
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Crystal Structure of Sars-Cov-2 Main Protease in Complex with Cpd-29 within 5.0Å range:
|
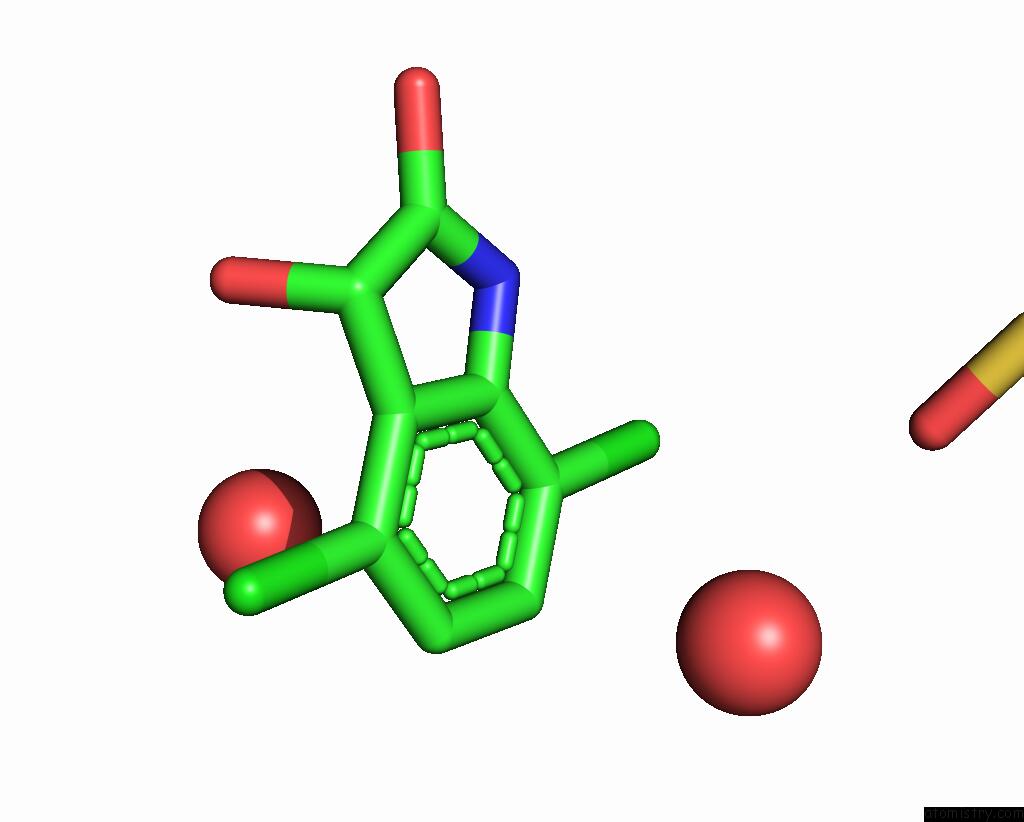
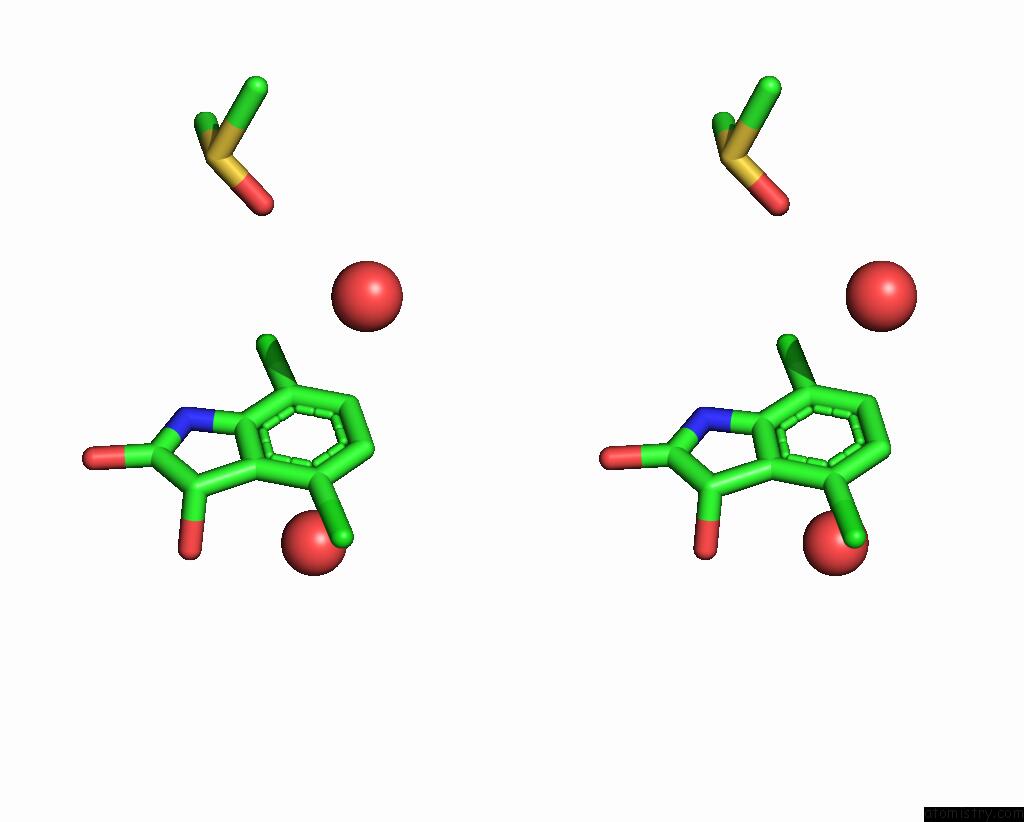
Chlorine binding site 3 out of 6 in 7gs6
Go back to
Chlorine binding site 3 out
of 6 in the Crystal Structure of Sars-Cov-2 Main Protease in Complex with Cpd-29
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Crystal Structure of Sars-Cov-2 Main Protease in Complex with Cpd-29 within 5.0Å range:
|
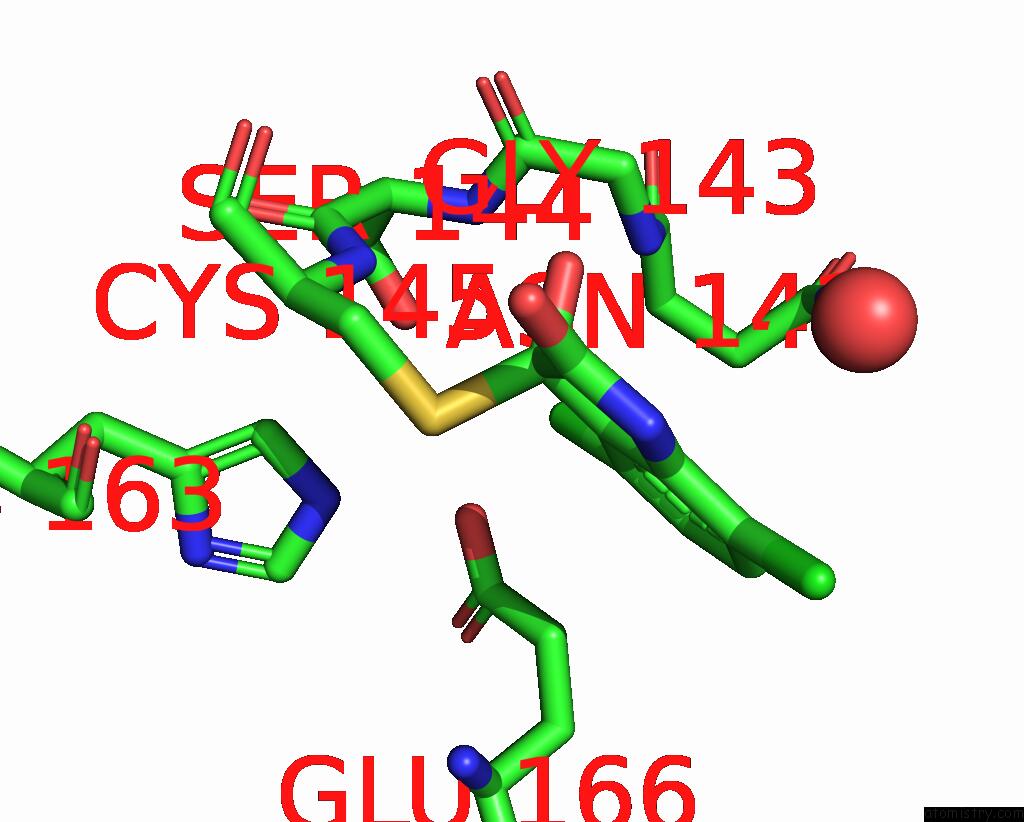
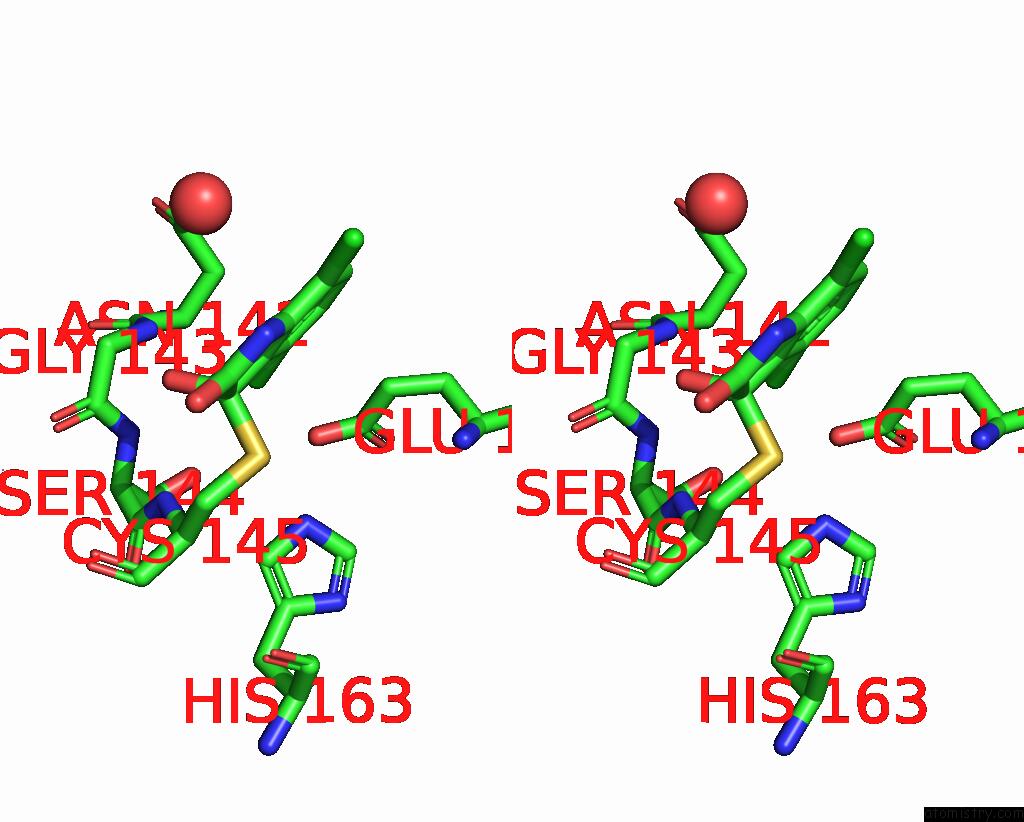
Chlorine binding site 4 out of 6 in 7gs6
Go back to
Chlorine binding site 4 out
of 6 in the Crystal Structure of Sars-Cov-2 Main Protease in Complex with Cpd-29
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of Crystal Structure of Sars-Cov-2 Main Protease in Complex with Cpd-29 within 5.0Å range:
|
Chlorine binding site 5 out of 6 in 7gs6
Go back to
Chlorine binding site 5 out
of 6 in the Crystal Structure of Sars-Cov-2 Main Protease in Complex with Cpd-29
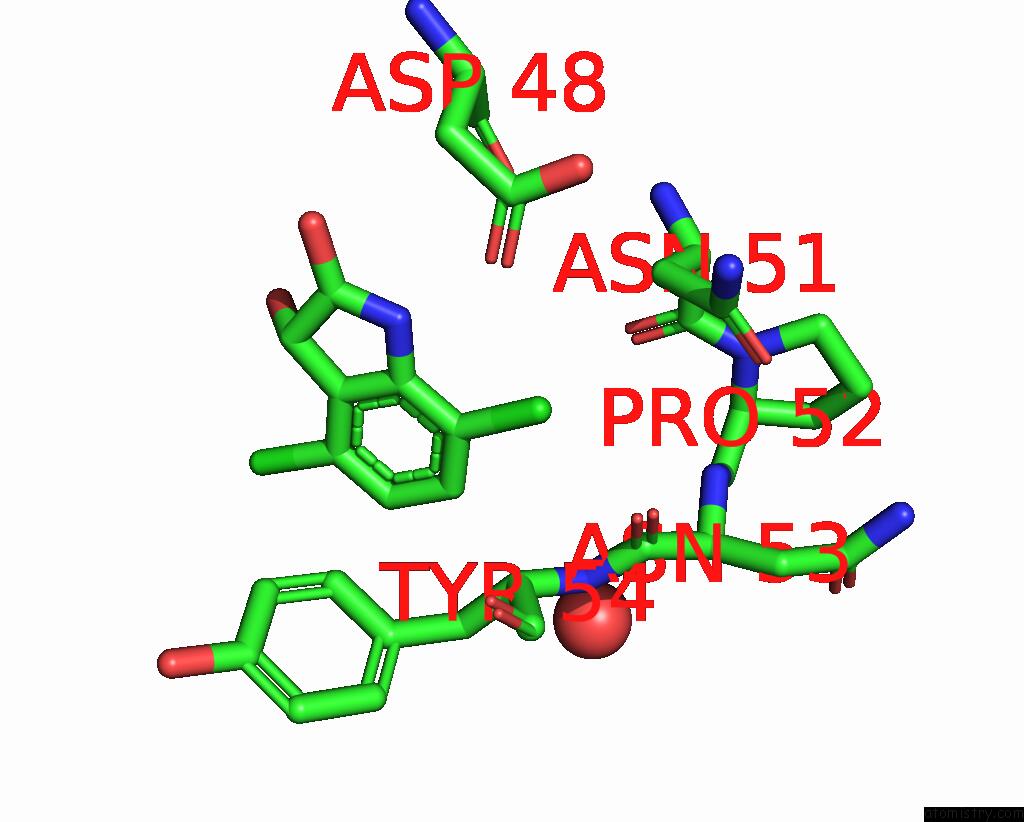
Mono view
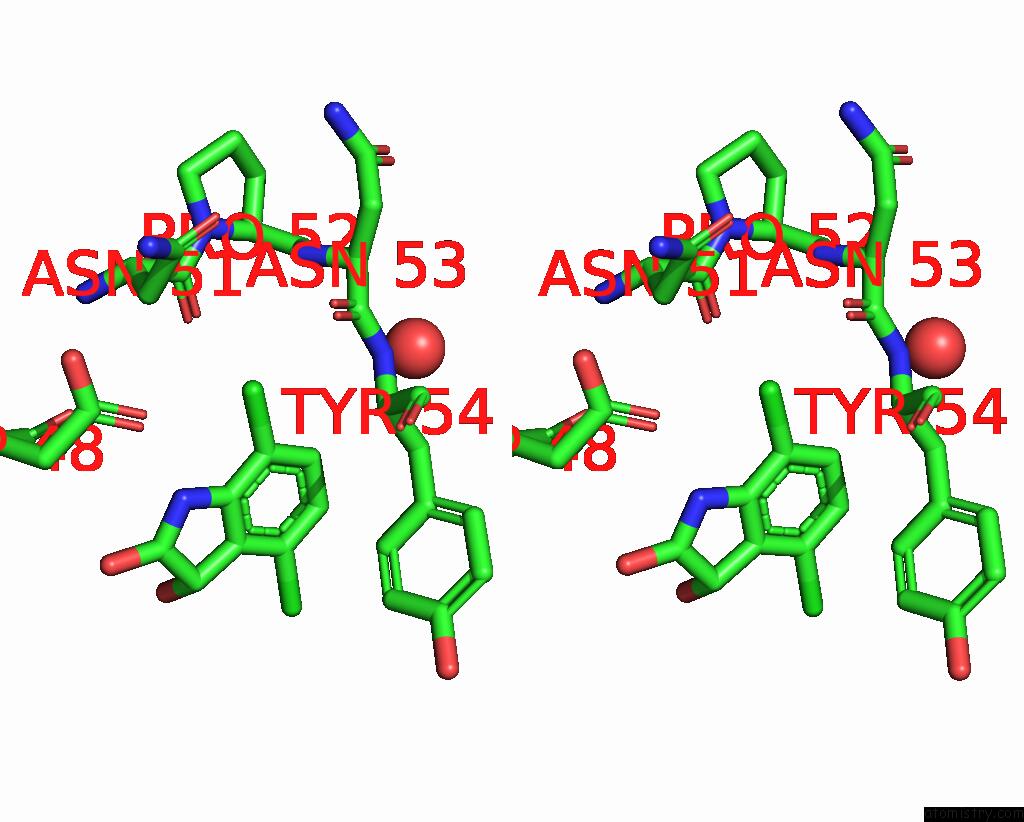
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 5 of Crystal Structure of Sars-Cov-2 Main Protease in Complex with Cpd-29 within 5.0Å range:
|
Chlorine binding site 6 out of 6 in 7gs6
Go back to
Chlorine binding site 6 out
of 6 in the Crystal Structure of Sars-Cov-2 Main Protease in Complex with Cpd-29
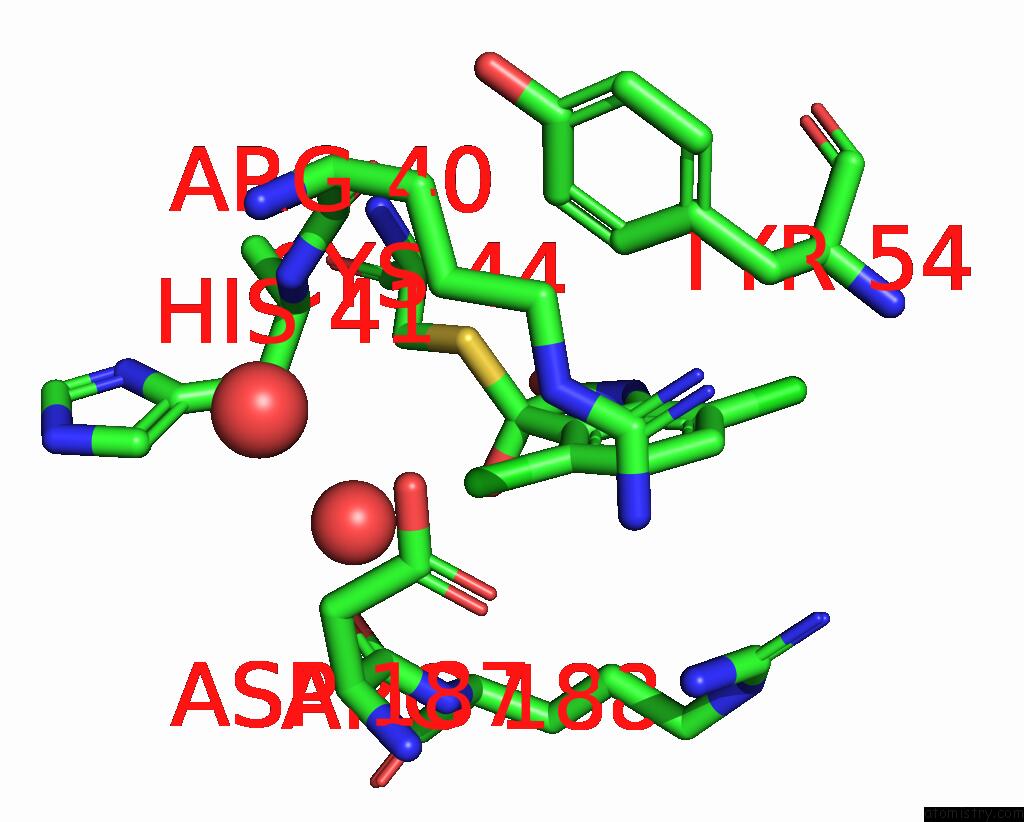
Mono view
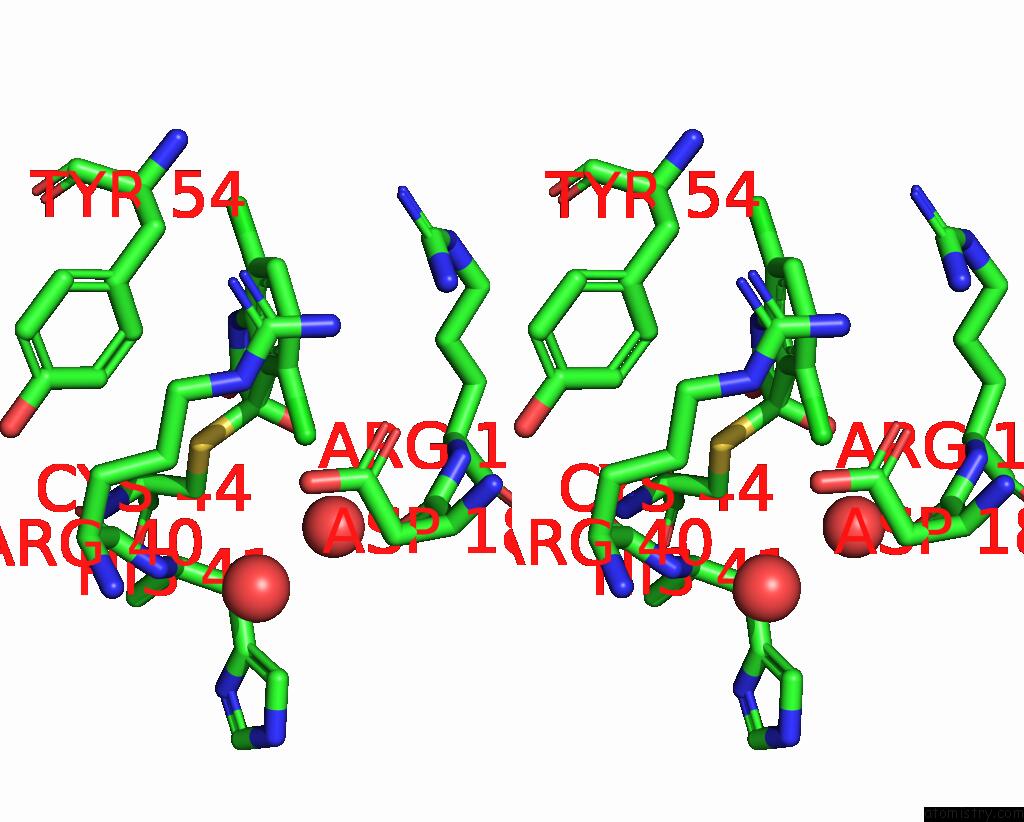
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 6 of Crystal Structure of Sars-Cov-2 Main Protease in Complex with Cpd-29 within 5.0Å range:
|
Reference:
C.Y.Huang,
A.Metz,
R.Lange,
N.Artico,
C.Potot,
J.Hazemann,
M.Muller,
M.Dos Santos,
A.Chambovey,
D.Ritz,
D.Eris,
S.Meyer,
G.Bourquin,
M.Sharpe,
A.Mac Sweeney.
Fragment-Based Screening Targeting An Open Form of the Sars-Cov-2 Main Protease Binding Pocket. Acta Crystallogr D Struct V. 80 123 2024BIOL.
ISSN: ISSN 2059-7983
PubMed: 38289714
DOI: 10.1107/S2059798324000329
Page generated: Sun Jul 13 02:06:06 2025
ISSN: ISSN 2059-7983
PubMed: 38289714
DOI: 10.1107/S2059798324000329
Last articles
K in 5UXEK in 5UZE
K in 5UUV
K in 5URS
K in 5URQ
K in 5UQH
K in 5UQG
K in 5UQF
K in 5UN7
K in 5UN2