Chlorine »
PDB 7hsq-7jho »
7i2m »
Chlorine in PDB 7i2m: Group Deposition For Crystallographic Fragment Screening of the NS5 Rna-Dependent Rna Polymerase From Dengue Virus Serotype 2 -- Crystal Structure of the NS5 Rna-Dependent Rna Polymerase From Dengue Virus Serotype 2 in Complex with Z1639162606 (DNV2_NS5A-X0567)
Protein crystallography data
The structure of Group Deposition For Crystallographic Fragment Screening of the NS5 Rna-Dependent Rna Polymerase From Dengue Virus Serotype 2 -- Crystal Structure of the NS5 Rna-Dependent Rna Polymerase From Dengue Virus Serotype 2 in Complex with Z1639162606 (DNV2_NS5A-X0567), PDB code: 7i2m
was solved by
J.C.Aschenbrenner,
M.Saini,
A.Chopra,
P.G.Marples,
B.H.Balcomb,
R.M.Lithgo,
D.Fearon,
F.Von Delft,
F.X.Ruiz,
E.Arnold,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 73.55 / 1.78 |
| Space group | I 2 2 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 82.364, 115.808, 147.054, 90, 90, 90 |
| R / Rfree (%) | 19.6 / 23.5 |
Other elements in 7i2m:
The structure of Group Deposition For Crystallographic Fragment Screening of the NS5 Rna-Dependent Rna Polymerase From Dengue Virus Serotype 2 -- Crystal Structure of the NS5 Rna-Dependent Rna Polymerase From Dengue Virus Serotype 2 in Complex with Z1639162606 (DNV2_NS5A-X0567) also contains other interesting chemical elements:
| Fluorine | (F) | 1 atom |
| Zinc | (Zn) | 2 atoms |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Group Deposition For Crystallographic Fragment Screening of the NS5 Rna-Dependent Rna Polymerase From Dengue Virus Serotype 2 -- Crystal Structure of the NS5 Rna-Dependent Rna Polymerase From Dengue Virus Serotype 2 in Complex with Z1639162606 (DNV2_NS5A-X0567)
(pdb code 7i2m). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total only one binding site of Chlorine was determined in the Group Deposition For Crystallographic Fragment Screening of the NS5 Rna-Dependent Rna Polymerase From Dengue Virus Serotype 2 -- Crystal Structure of the NS5 Rna-Dependent Rna Polymerase From Dengue Virus Serotype 2 in Complex with Z1639162606 (DNV2_NS5A-X0567), PDB code: 7i2m:
In total only one binding site of Chlorine was determined in the Group Deposition For Crystallographic Fragment Screening of the NS5 Rna-Dependent Rna Polymerase From Dengue Virus Serotype 2 -- Crystal Structure of the NS5 Rna-Dependent Rna Polymerase From Dengue Virus Serotype 2 in Complex with Z1639162606 (DNV2_NS5A-X0567), PDB code: 7i2m:
Chlorine binding site 1 out of 1 in 7i2m
Go back to
Chlorine binding site 1 out
of 1 in the Group Deposition For Crystallographic Fragment Screening of the NS5 Rna-Dependent Rna Polymerase From Dengue Virus Serotype 2 -- Crystal Structure of the NS5 Rna-Dependent Rna Polymerase From Dengue Virus Serotype 2 in Complex with Z1639162606 (DNV2_NS5A-X0567)
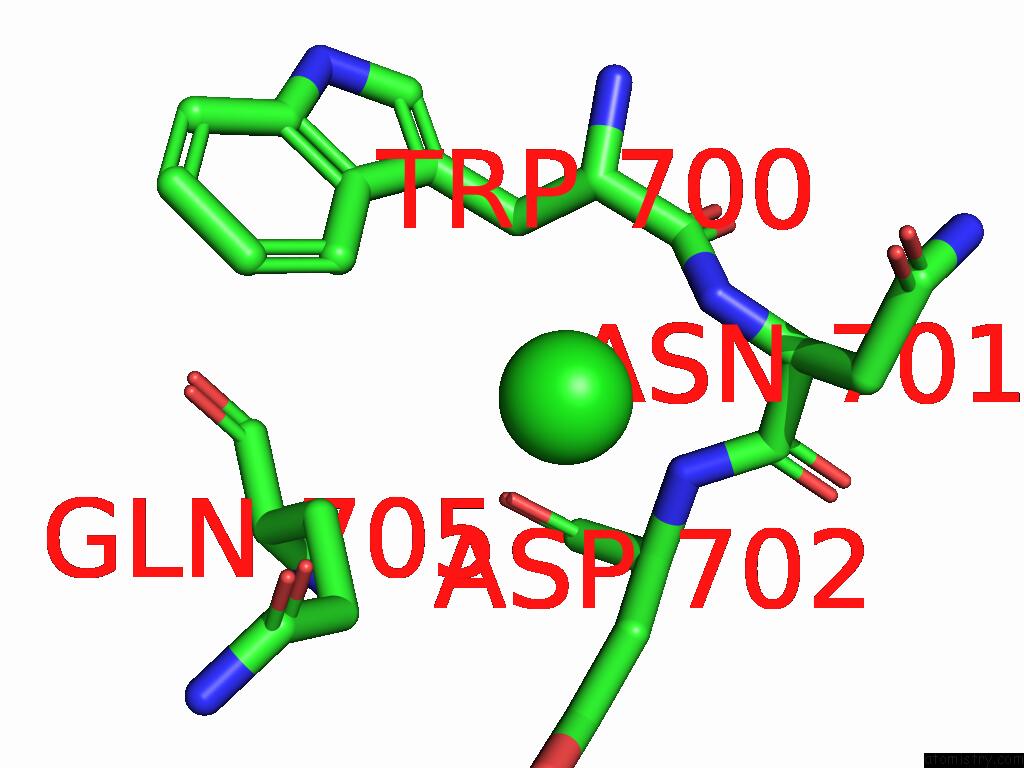
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Group Deposition For Crystallographic Fragment Screening of the NS5 Rna-Dependent Rna Polymerase From Dengue Virus Serotype 2 -- Crystal Structure of the NS5 Rna-Dependent Rna Polymerase From Dengue Virus Serotype 2 in Complex with Z1639162606 (DNV2_NS5A-X0567) within 5.0Å range:
|
Reference:
J.C.Aschenbrenner,
M.Saini,
A.Chopra,
P.G.Marples,
B.H.Balcomb,
R.M.Lithgo,
D.Fearon,
F.Von Delft,
F.X.Ruiz,
E.Arnold.
Group Deposition For Crystallographic Fragment Screening of the NS5 Rna-Dependent Rna Polymerase From Dengue Virus Serotype 2 To Be Published.
Page generated: Sun Jul 13 02:55:21 2025
Last articles
Mn in 9LJUMn in 9LJW
Mn in 9LJS
Mn in 9LJR
Mn in 9LJT
Mn in 9LJV
Mg in 9UA2
Mg in 9R96
Mg in 9VM1
Mg in 9P01