Chlorine »
PDB 7l1g-7l9j »
7l3f »
Chlorine in PDB 7l3f: T4 Lysozyme L99A - 4-Iodotoluene - Rt
Enzymatic activity of T4 Lysozyme L99A - 4-Iodotoluene - Rt
All present enzymatic activity of T4 Lysozyme L99A - 4-Iodotoluene - Rt:
3.2.1.17;
3.2.1.17;
Protein crystallography data
The structure of T4 Lysozyme L99A - 4-Iodotoluene - Rt, PDB code: 7l3f
was solved by
M.Fischer,
S.Y.C.Bradford,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 35.93 / 1.49 |
| Space group | P 32 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 61.129, 61.129, 97.868, 90, 90, 120 |
| R / Rfree (%) | 14.8 / 17 |
Other elements in 7l3f:
The structure of T4 Lysozyme L99A - 4-Iodotoluene - Rt also contains other interesting chemical elements:
| Iodine | (I) | 2 atoms |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the T4 Lysozyme L99A - 4-Iodotoluene - Rt
(pdb code 7l3f). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total only one binding site of Chlorine was determined in the T4 Lysozyme L99A - 4-Iodotoluene - Rt, PDB code: 7l3f:
In total only one binding site of Chlorine was determined in the T4 Lysozyme L99A - 4-Iodotoluene - Rt, PDB code: 7l3f:
Chlorine binding site 1 out of 1 in 7l3f
Go back to
Chlorine binding site 1 out
of 1 in the T4 Lysozyme L99A - 4-Iodotoluene - Rt
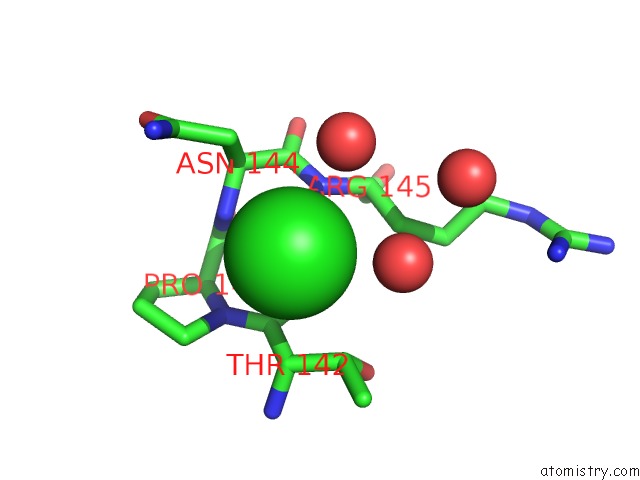
Mono view
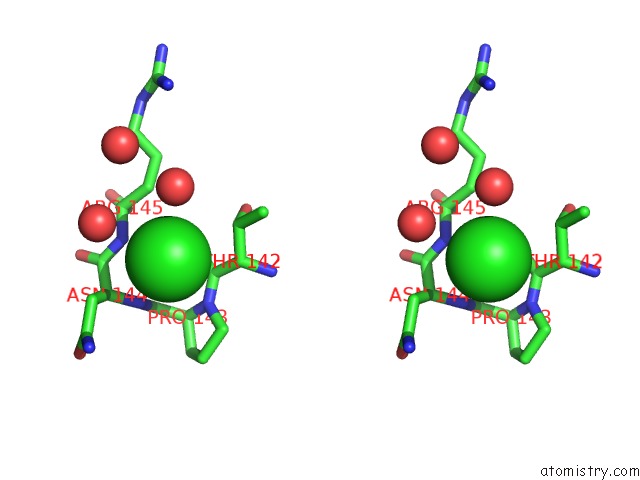
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of T4 Lysozyme L99A - 4-Iodotoluene - Rt within 5.0Å range:
|
Reference:
S.Y.C.Bradford,
L.El Khoury,
Y.Ge,
M.Osato,
D.L.Mobley,
M.Fischer.
Temperature Artifacts in Protein Structures Bias Ligand-Binding Predictions. Chem Sci V. 12 11275 2021.
ISSN: ISSN 2041-6520
PubMed: 34667539
DOI: 10.1039/D1SC02751D
Page generated: Sun Jul 13 03:33:54 2025
ISSN: ISSN 2041-6520
PubMed: 34667539
DOI: 10.1039/D1SC02751D
Last articles
I in 3NUJI in 3OHF
I in 3O2E
I in 3O8H
I in 3NU9
I in 3MEN
I in 3NTE
I in 3NJB
I in 3MXM
I in 3NSJ