Chlorine »
PDB 7oke-7op6 »
7ood »
Chlorine in PDB 7ood: Mycoplasma Pneumoniae 50S Subunit of Ribosomes in Chloramphenicol- Treated Cells
Other elements in 7ood:
The structure of Mycoplasma Pneumoniae 50S Subunit of Ribosomes in Chloramphenicol- Treated Cells also contains other interesting chemical elements:
| Zinc | (Zn) | 3 atoms |
| Magnesium | (Mg) | 25 atoms |
| Potassium | (K) | 1 atom |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Mycoplasma Pneumoniae 50S Subunit of Ribosomes in Chloramphenicol- Treated Cells
(pdb code 7ood). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 2 binding sites of Chlorine where determined in the Mycoplasma Pneumoniae 50S Subunit of Ribosomes in Chloramphenicol- Treated Cells, PDB code: 7ood:
Jump to Chlorine binding site number: 1; 2;
In total 2 binding sites of Chlorine where determined in the Mycoplasma Pneumoniae 50S Subunit of Ribosomes in Chloramphenicol- Treated Cells, PDB code: 7ood:
Jump to Chlorine binding site number: 1; 2;
Chlorine binding site 1 out of 2 in 7ood
Go back to
Chlorine binding site 1 out
of 2 in the Mycoplasma Pneumoniae 50S Subunit of Ribosomes in Chloramphenicol- Treated Cells
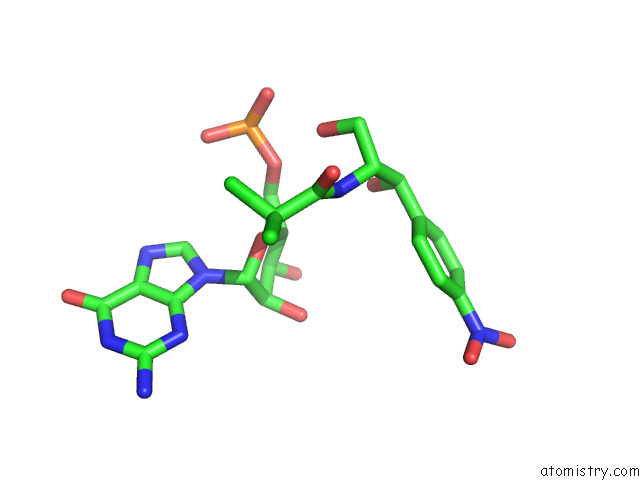
Mono view
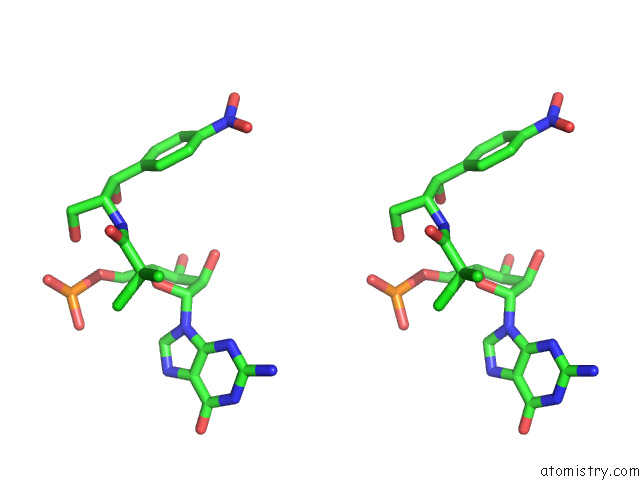
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Mycoplasma Pneumoniae 50S Subunit of Ribosomes in Chloramphenicol- Treated Cells within 5.0Å range:
|
Chlorine binding site 2 out of 2 in 7ood
Go back to
Chlorine binding site 2 out
of 2 in the Mycoplasma Pneumoniae 50S Subunit of Ribosomes in Chloramphenicol- Treated Cells
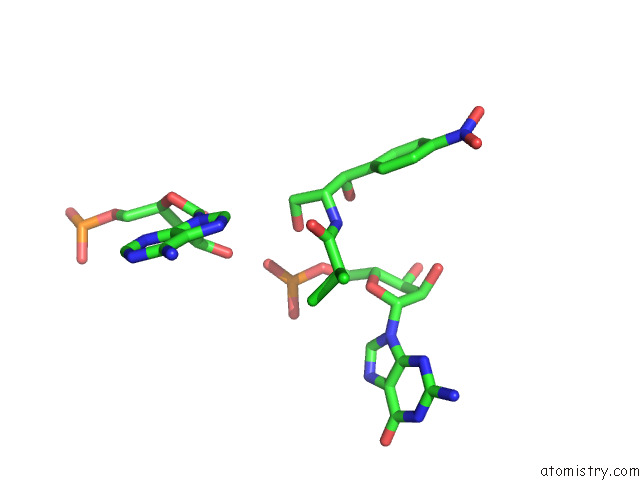
Mono view
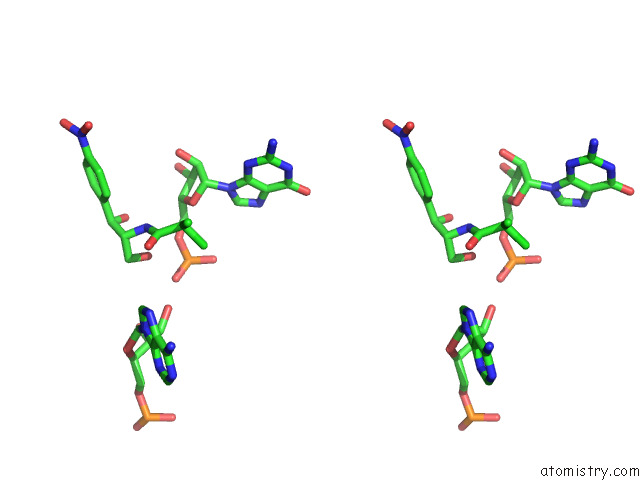
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Mycoplasma Pneumoniae 50S Subunit of Ribosomes in Chloramphenicol- Treated Cells within 5.0Å range:
|
Reference:
L.Xue,
S.Lenz,
M.Zimmermann-Kogadeeva,
D.Tegunov,
P.Cramer,
P.Bork,
J.Rappsilber,
J.Mahamid.
Visualizing Translation Dynamics at Atomic Detail Inside A Bacterial Cell. Nature V. 610 205 2022.
ISSN: ESSN 1476-4687
PubMed: 36171285
DOI: 10.1038/S41586-022-05255-2
Page generated: Sun Jul 13 05:18:35 2025
ISSN: ESSN 1476-4687
PubMed: 36171285
DOI: 10.1038/S41586-022-05255-2
Last articles
Mg in 4HGNMg in 4HGP
Mg in 4HE2
Mg in 4HEC
Mg in 4HE0
Mg in 4HEO
Mg in 4HE1
Mg in 4HCL
Mg in 4HDQ
Mg in 4HCJ