Chlorine »
PDB 7pxl-7q1o »
7q0i »
Chlorine in PDB 7q0i: Crystal Structure of the N-Terminal Domain of Sars-Cov-2 Beta Variant Spike Glycoprotein in Complex with Beta-43
Protein crystallography data
The structure of Crystal Structure of the N-Terminal Domain of Sars-Cov-2 Beta Variant Spike Glycoprotein in Complex with Beta-43, PDB code: 7q0i
was solved by
D.Zhou,
J.Ren,
D.I.Stuart,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 143.10 / 2.39 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 117.83, 66.964, 143.238, 90, 92.54, 90 |
| R / Rfree (%) | 20.2 / 24.2 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Crystal Structure of the N-Terminal Domain of Sars-Cov-2 Beta Variant Spike Glycoprotein in Complex with Beta-43
(pdb code 7q0i). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 6 binding sites of Chlorine where determined in the Crystal Structure of the N-Terminal Domain of Sars-Cov-2 Beta Variant Spike Glycoprotein in Complex with Beta-43, PDB code: 7q0i:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5; 6;
In total 6 binding sites of Chlorine where determined in the Crystal Structure of the N-Terminal Domain of Sars-Cov-2 Beta Variant Spike Glycoprotein in Complex with Beta-43, PDB code: 7q0i:
Jump to Chlorine binding site number: 1; 2; 3; 4; 5; 6;
Chlorine binding site 1 out of 6 in 7q0i
Go back to
Chlorine binding site 1 out
of 6 in the Crystal Structure of the N-Terminal Domain of Sars-Cov-2 Beta Variant Spike Glycoprotein in Complex with Beta-43
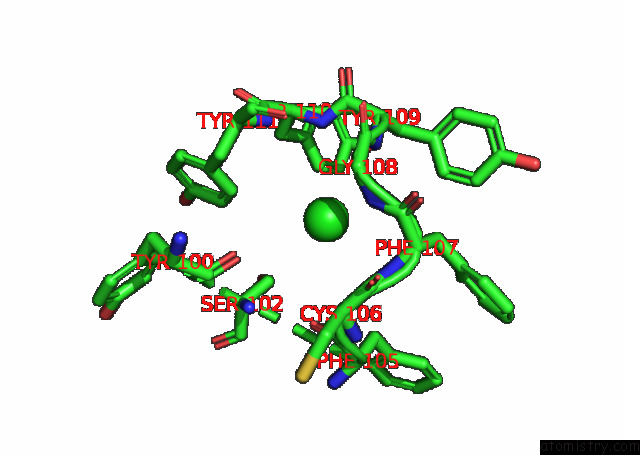
Mono view
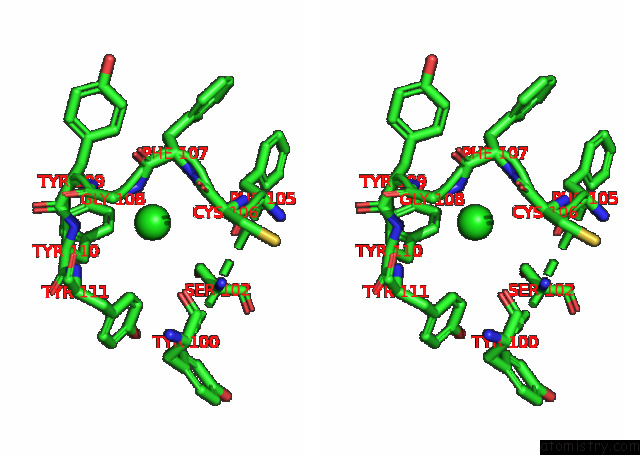
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Crystal Structure of the N-Terminal Domain of Sars-Cov-2 Beta Variant Spike Glycoprotein in Complex with Beta-43 within 5.0Å range:
|
Chlorine binding site 2 out of 6 in 7q0i
Go back to
Chlorine binding site 2 out
of 6 in the Crystal Structure of the N-Terminal Domain of Sars-Cov-2 Beta Variant Spike Glycoprotein in Complex with Beta-43
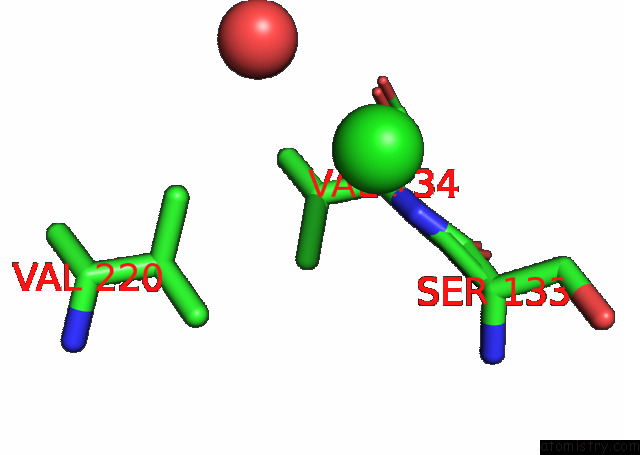
Mono view
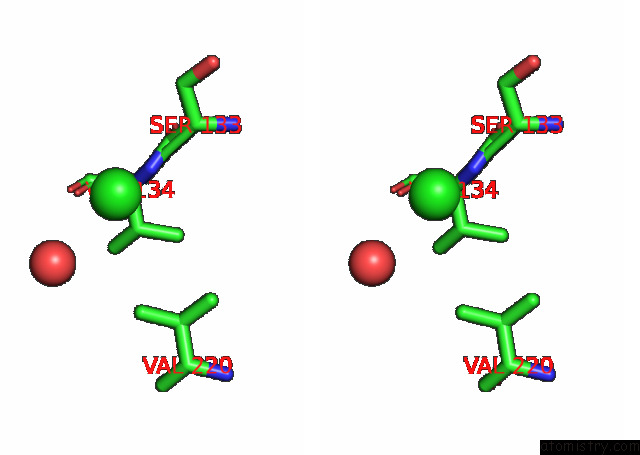
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Crystal Structure of the N-Terminal Domain of Sars-Cov-2 Beta Variant Spike Glycoprotein in Complex with Beta-43 within 5.0Å range:
|
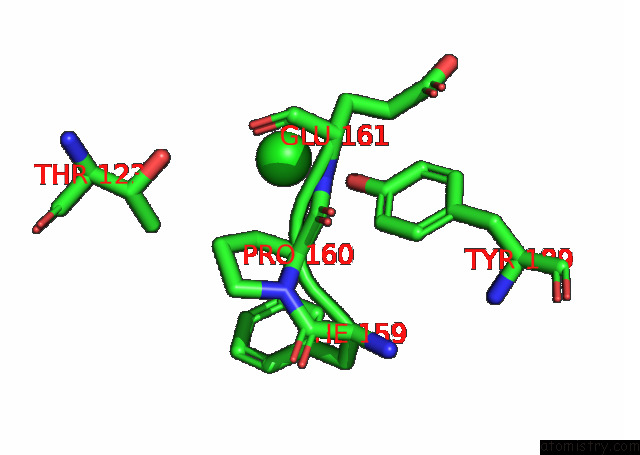
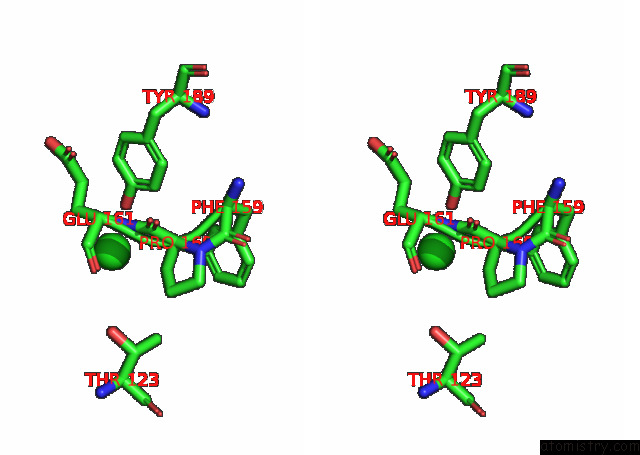
Chlorine binding site 3 out of 6 in 7q0i
Go back to
Chlorine binding site 3 out
of 6 in the Crystal Structure of the N-Terminal Domain of Sars-Cov-2 Beta Variant Spike Glycoprotein in Complex with Beta-43
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Crystal Structure of the N-Terminal Domain of Sars-Cov-2 Beta Variant Spike Glycoprotein in Complex with Beta-43 within 5.0Å range:
|
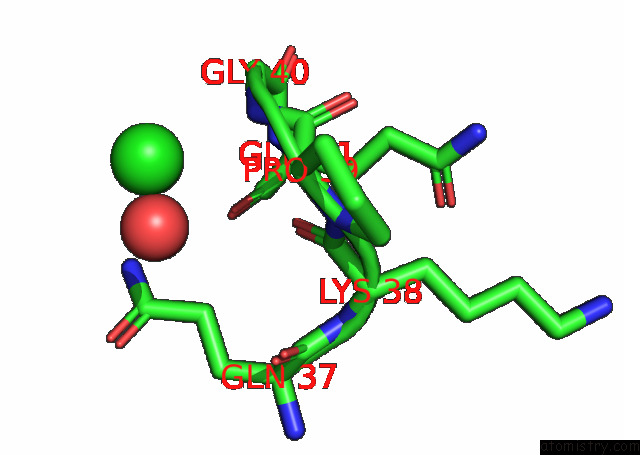
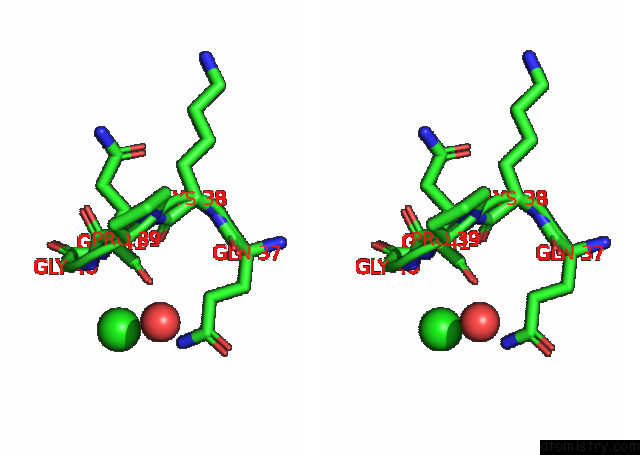
Chlorine binding site 4 out of 6 in 7q0i
Go back to
Chlorine binding site 4 out
of 6 in the Crystal Structure of the N-Terminal Domain of Sars-Cov-2 Beta Variant Spike Glycoprotein in Complex with Beta-43
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of Crystal Structure of the N-Terminal Domain of Sars-Cov-2 Beta Variant Spike Glycoprotein in Complex with Beta-43 within 5.0Å range:
|
Chlorine binding site 5 out of 6 in 7q0i
Go back to
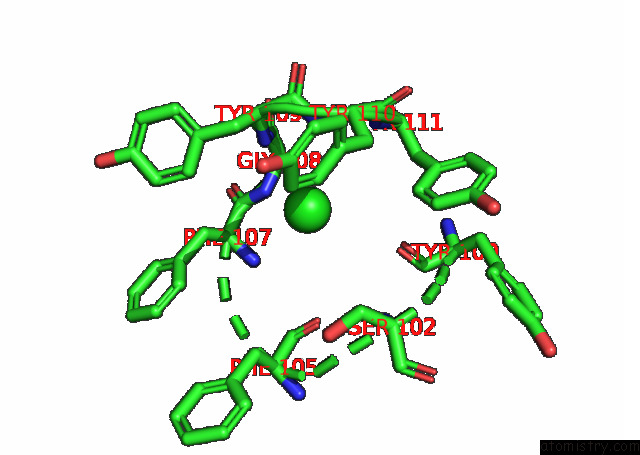
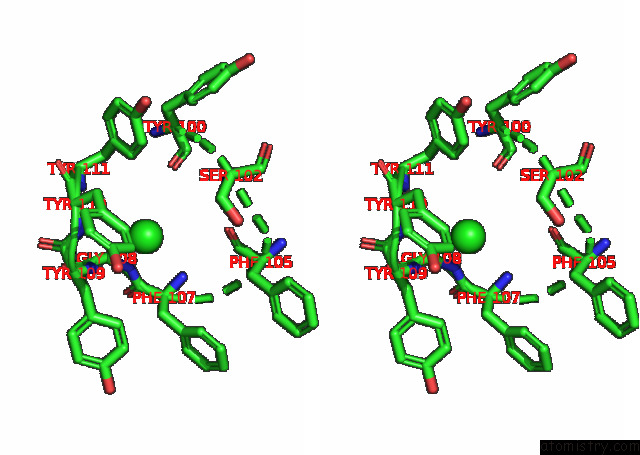
Chlorine binding site 5 out
of 6 in the Crystal Structure of the N-Terminal Domain of Sars-Cov-2 Beta Variant Spike Glycoprotein in Complex with Beta-43
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 5 of Crystal Structure of the N-Terminal Domain of Sars-Cov-2 Beta Variant Spike Glycoprotein in Complex with Beta-43 within 5.0Å range:
|
Chlorine binding site 6 out of 6 in 7q0i
Go back to
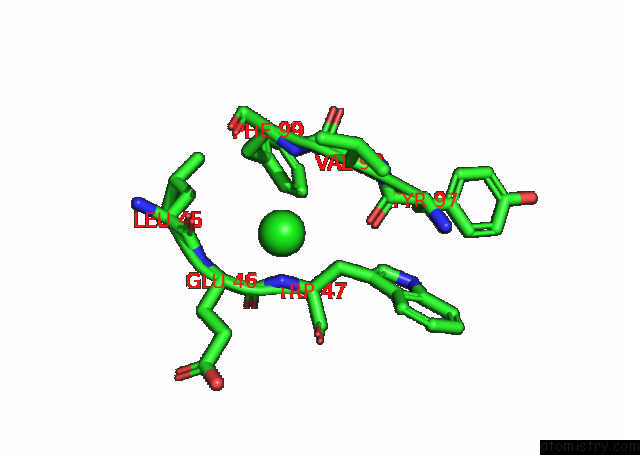
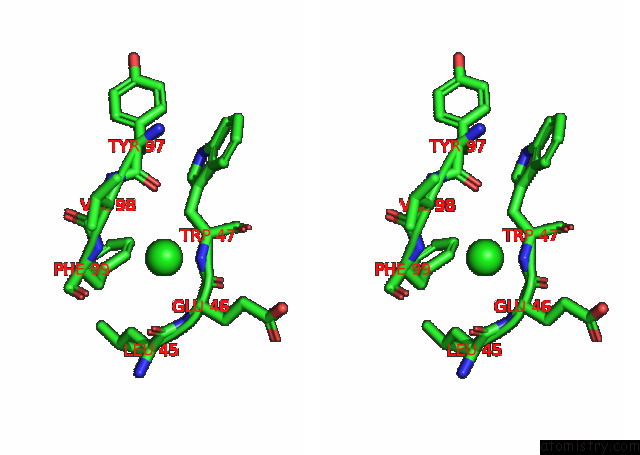
Chlorine binding site 6 out
of 6 in the Crystal Structure of the N-Terminal Domain of Sars-Cov-2 Beta Variant Spike Glycoprotein in Complex with Beta-43
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 6 of Crystal Structure of the N-Terminal Domain of Sars-Cov-2 Beta Variant Spike Glycoprotein in Complex with Beta-43 within 5.0Å range:
|
Reference:
C.Liu,
D.Zhou,
R.Nutalai,
H.M.Duyvesteyn,
A.Tuekprakhon,
H.M.Ginn,
W.Dejnirattisai,
P.Supasa,
A.J.Mentzer,
B.Wang,
J.B.Case,
Y.Zhao,
D.T.Skelly,
R.E.Chen,
S.A.Johnson,
T.G.Ritter,
C.Mason,
T.Malik,
N.Temperton,
N.G.Paterson,
M.A.Williams,
D.R.Hall,
D.K.Clare,
A.Howe,
P.J.Goulder,
E.E.Fry,
M.S.Diamond,
J.Mongkolsapaya,
J.Ren,
D.I.Stuart,
G.R.Screaton.
The Antibody Response to Sars-Cov-2 Beta Underscores the Antigenic Distance to Other Variants Cell Host Microbe 2021.
ISSN: ESSN 1934-6069
DOI: 10.1016/J.CHOM.2021.11.013
Page generated: Sun Jul 13 06:04:16 2025
ISSN: ESSN 1934-6069
DOI: 10.1016/J.CHOM.2021.11.013
Last articles
K in 8CGIK in 8CG1
K in 8CFY
K in 8CG2
K in 8CFX
K in 8CG0
K in 8CFW
K in 8CFV
K in 8CFU
K in 8CFT