Chlorine »
PDB 7qip-7qzr »
7qqc »
Chlorine in PDB 7qqc: Structure of Ctx-M-15 K73A Mutant
Enzymatic activity of Structure of Ctx-M-15 K73A Mutant
All present enzymatic activity of Structure of Ctx-M-15 K73A Mutant:
3.5.2.6;
3.5.2.6;
Protein crystallography data
The structure of Structure of Ctx-M-15 K73A Mutant, PDB code: 7qqc
was solved by
C.L.Tooke,
P.Hinchliffe,
J.Spencer,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 35.91 / 0.95 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 44.896, 45.552, 116.705, 90, 90, 90 |
| R / Rfree (%) | 14 / 15.6 |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Structure of Ctx-M-15 K73A Mutant
(pdb code 7qqc). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total only one binding site of Chlorine was determined in the Structure of Ctx-M-15 K73A Mutant, PDB code: 7qqc:
In total only one binding site of Chlorine was determined in the Structure of Ctx-M-15 K73A Mutant, PDB code: 7qqc:
Chlorine binding site 1 out of 1 in 7qqc
Go back to
Chlorine binding site 1 out
of 1 in the Structure of Ctx-M-15 K73A Mutant
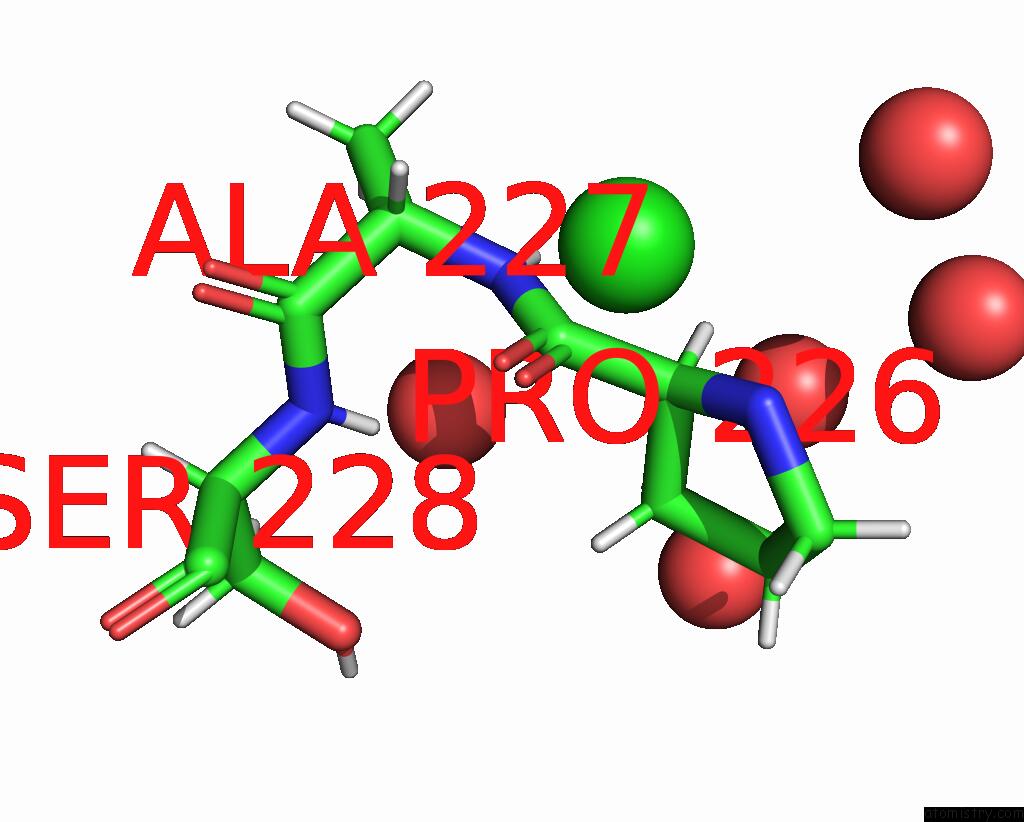
Mono view
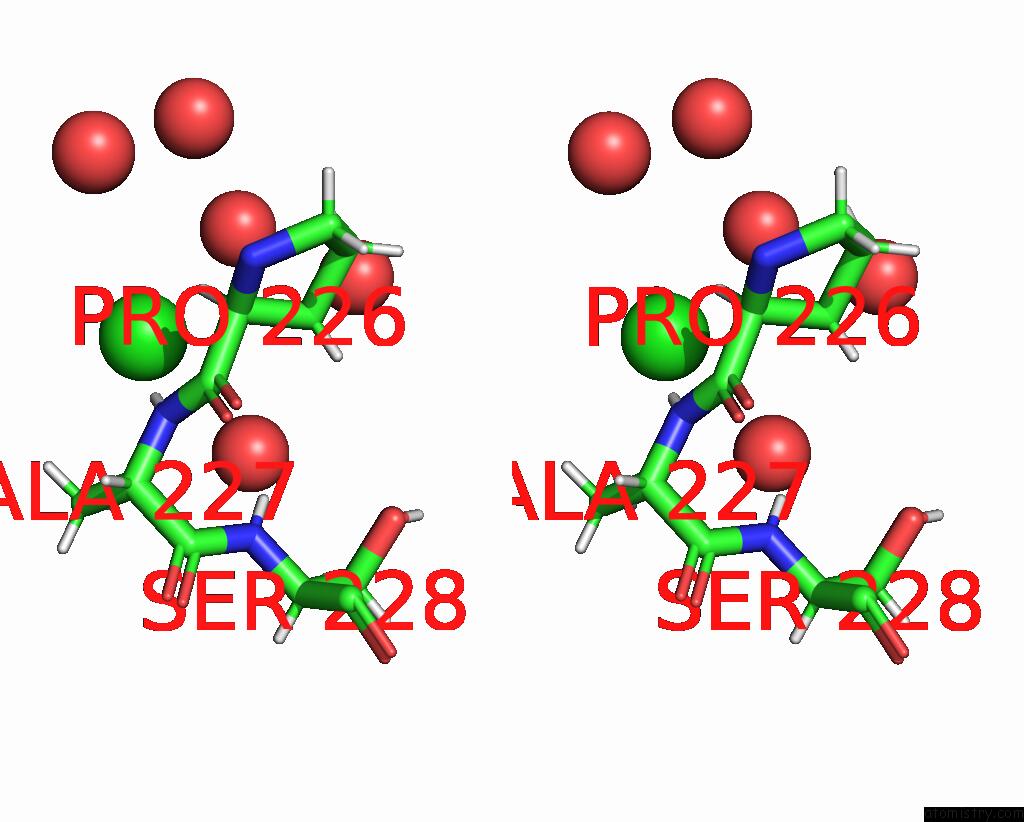
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Structure of Ctx-M-15 K73A Mutant within 5.0Å range:
|
Reference:
P.Hinchliffe,
C.L.Tooke,
C.R.Bethel,
B.Wang,
C.Arthur,
K.J.Heesom,
S.Shapiro,
D.M.Schlatzer,
K.M.Papp-Wallace,
R.A.Bonomo,
J.Spencer.
Penicillanic Acid Sulfones Inactivate the Extended-Spectrum Beta-Lactamase Ctx-M-15 Through Formation of A Serine-Lysine Cross-Link: An Alternative Mechanism of Beta-Lactamase Inhibition. Mbio V. 13 79321 2022.
ISSN: ESSN 2150-7511
PubMed: 35612361
DOI: 10.1128/MBIO.01793-21
Page generated: Sun Jul 13 06:26:26 2025
ISSN: ESSN 2150-7511
PubMed: 35612361
DOI: 10.1128/MBIO.01793-21
Last articles
Mg in 1W5FMg in 1W5A
Mg in 1W58
Mg in 1W2B
Mg in 1VQP
Mg in 1VQO
Mg in 1W55
Mg in 1W54
Mg in 1W4B
Mg in 1W49