Chlorine »
PDB 7r97-7rjj »
7reg »
Chlorine in PDB 7reg: DFRA1 Complexed with Nadph and 4'-Chloro-3'-(4-(2,4-Diamino-6- Ethylpyrimidin-5-Yl)But-3-Yn-2-Yl)-[1,1'-Biphenyl]-4-Carboxamide (UCP1228)
Enzymatic activity of DFRA1 Complexed with Nadph and 4'-Chloro-3'-(4-(2,4-Diamino-6- Ethylpyrimidin-5-Yl)But-3-Yn-2-Yl)-[1,1'-Biphenyl]-4-Carboxamide (UCP1228)
All present enzymatic activity of DFRA1 Complexed with Nadph and 4'-Chloro-3'-(4-(2,4-Diamino-6- Ethylpyrimidin-5-Yl)But-3-Yn-2-Yl)-[1,1'-Biphenyl]-4-Carboxamide (UCP1228):
1.5.1.3;
1.5.1.3;
Protein crystallography data
The structure of DFRA1 Complexed with Nadph and 4'-Chloro-3'-(4-(2,4-Diamino-6- Ethylpyrimidin-5-Yl)But-3-Yn-2-Yl)-[1,1'-Biphenyl]-4-Carboxamide (UCP1228), PDB code: 7reg
was solved by
M.N.Lombardo,
D.L.Wright,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 43.15 / 1.77 |
| Space group | P 31 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 71.86, 71.86, 119.781, 90, 90, 120 |
| R / Rfree (%) | 19 / 22.1 |
Other elements in 7reg:
The structure of DFRA1 Complexed with Nadph and 4'-Chloro-3'-(4-(2,4-Diamino-6- Ethylpyrimidin-5-Yl)But-3-Yn-2-Yl)-[1,1'-Biphenyl]-4-Carboxamide (UCP1228) also contains other interesting chemical elements:
| Calcium | (Ca) | 1 atom |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the DFRA1 Complexed with Nadph and 4'-Chloro-3'-(4-(2,4-Diamino-6- Ethylpyrimidin-5-Yl)But-3-Yn-2-Yl)-[1,1'-Biphenyl]-4-Carboxamide (UCP1228)
(pdb code 7reg). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 2 binding sites of Chlorine where determined in the DFRA1 Complexed with Nadph and 4'-Chloro-3'-(4-(2,4-Diamino-6- Ethylpyrimidin-5-Yl)But-3-Yn-2-Yl)-[1,1'-Biphenyl]-4-Carboxamide (UCP1228), PDB code: 7reg:
Jump to Chlorine binding site number: 1; 2;
In total 2 binding sites of Chlorine where determined in the DFRA1 Complexed with Nadph and 4'-Chloro-3'-(4-(2,4-Diamino-6- Ethylpyrimidin-5-Yl)But-3-Yn-2-Yl)-[1,1'-Biphenyl]-4-Carboxamide (UCP1228), PDB code: 7reg:
Jump to Chlorine binding site number: 1; 2;
Chlorine binding site 1 out of 2 in 7reg
Go back to
Chlorine binding site 1 out
of 2 in the DFRA1 Complexed with Nadph and 4'-Chloro-3'-(4-(2,4-Diamino-6- Ethylpyrimidin-5-Yl)But-3-Yn-2-Yl)-[1,1'-Biphenyl]-4-Carboxamide (UCP1228)
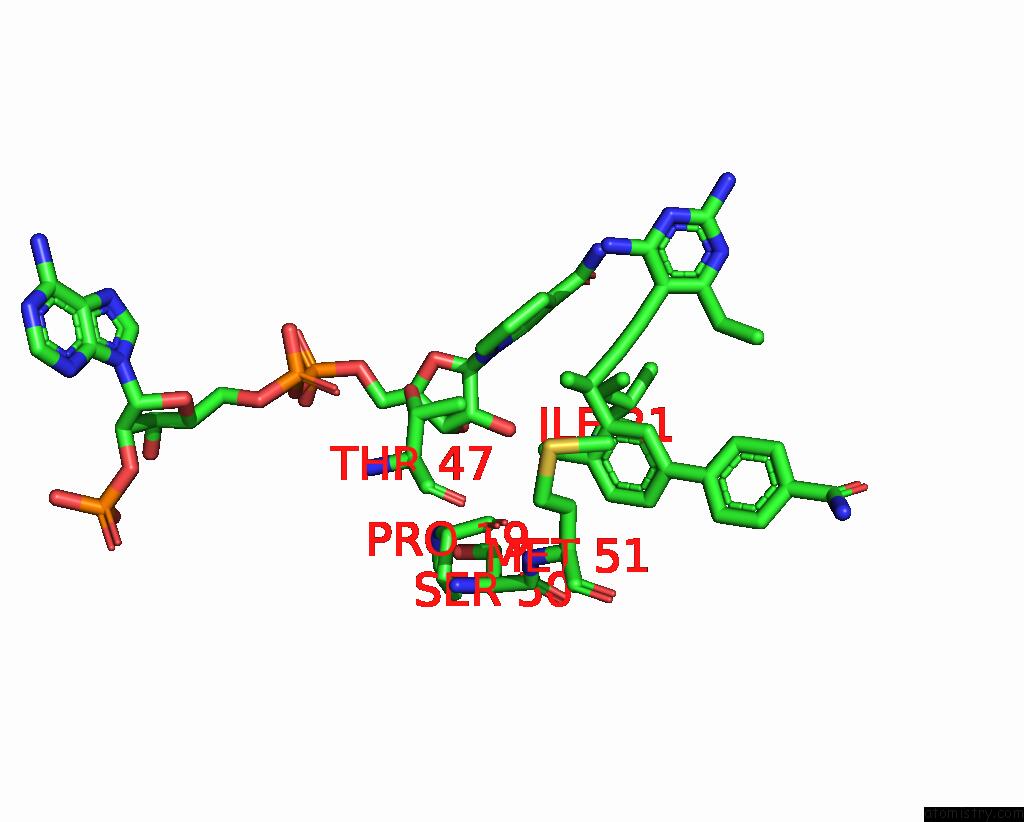
Mono view
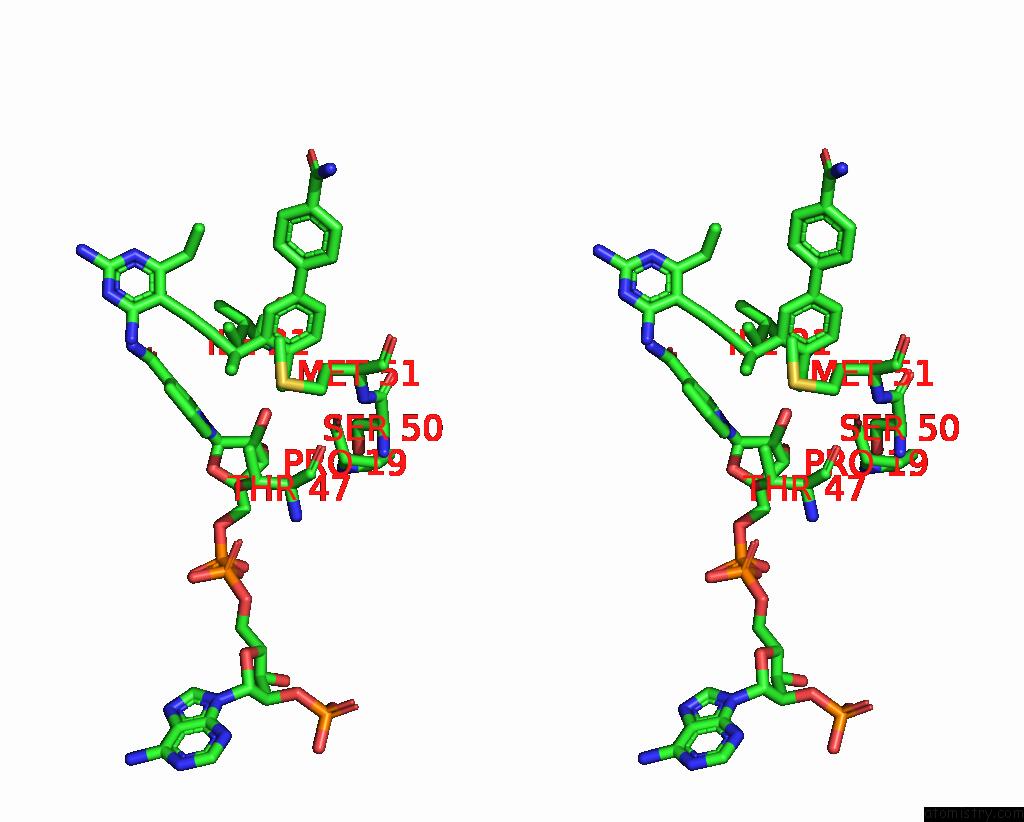
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of DFRA1 Complexed with Nadph and 4'-Chloro-3'-(4-(2,4-Diamino-6- Ethylpyrimidin-5-Yl)But-3-Yn-2-Yl)-[1,1'-Biphenyl]-4-Carboxamide (UCP1228) within 5.0Å range:
|
Chlorine binding site 2 out of 2 in 7reg
Go back to
Chlorine binding site 2 out
of 2 in the DFRA1 Complexed with Nadph and 4'-Chloro-3'-(4-(2,4-Diamino-6- Ethylpyrimidin-5-Yl)But-3-Yn-2-Yl)-[1,1'-Biphenyl]-4-Carboxamide (UCP1228)
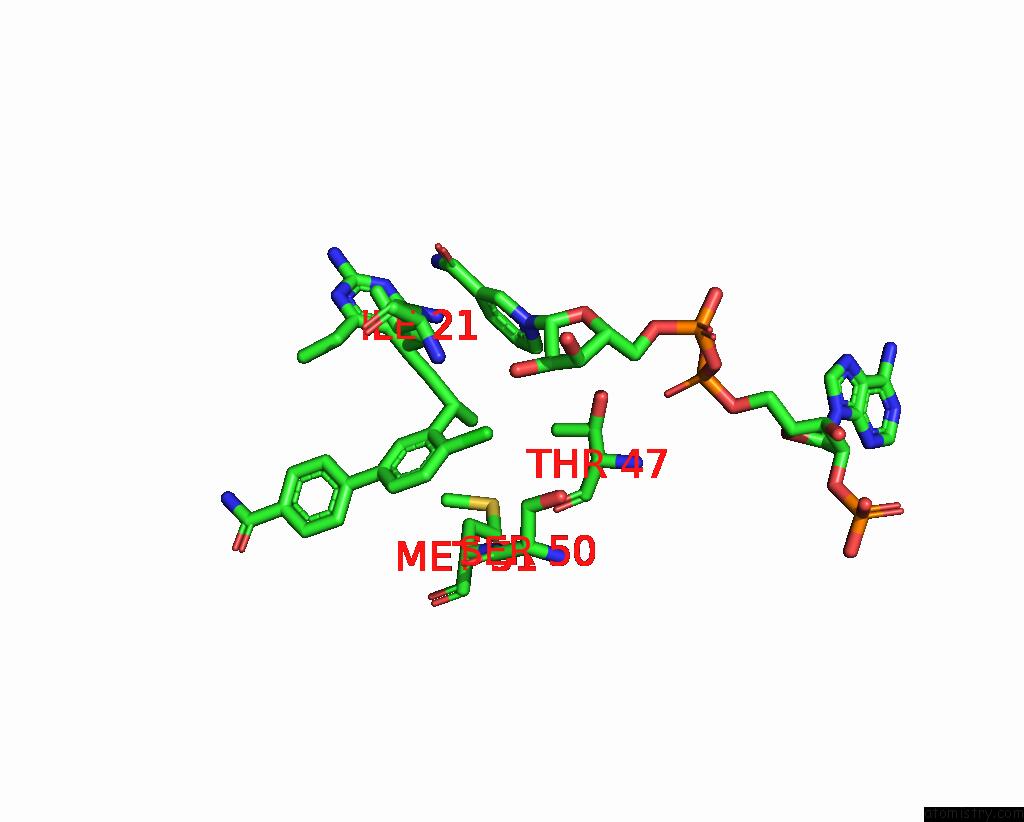
Mono view
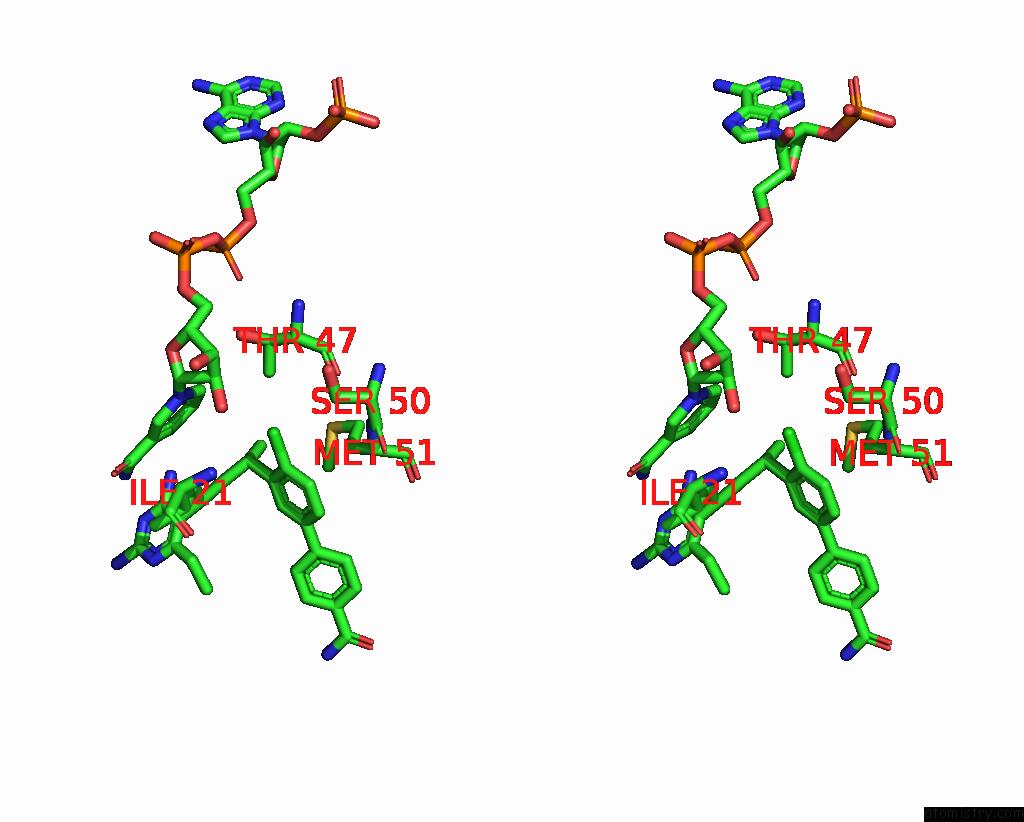
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of DFRA1 Complexed with Nadph and 4'-Chloro-3'-(4-(2,4-Diamino-6- Ethylpyrimidin-5-Yl)But-3-Yn-2-Yl)-[1,1'-Biphenyl]-4-Carboxamide (UCP1228) within 5.0Å range:
|
Reference:
J.Krucinska,
M.N.Lombardo,
H.Erlandsen,
A.Estrada,
D.Si,
K.Viswanathan,
D.L.Wright.
Structure-Guided Functional Studies of Plasmid-Encoded Dihydrofolate Reductases Reveal A Common Mechanism of Trimethoprim Resistance in Gram-Negative Pathogens. Commun Biol V. 5 459 2022.
ISSN: ESSN 2399-3642
PubMed: 35562546
DOI: 10.1038/S42003-022-03384-Y
Page generated: Sun Jul 13 06:36:58 2025
ISSN: ESSN 2399-3642
PubMed: 35562546
DOI: 10.1038/S42003-022-03384-Y
Last articles
K in 8CGJK in 8CGI
K in 8CG1
K in 8CFY
K in 8CG2
K in 8CFX
K in 8CG0
K in 8CFW
K in 8CFV
K in 8CFU