Chlorine »
PDB 7ti0-7tp6 »
7tn4 »
Chlorine in PDB 7tn4: Diphosphoinositol Polyphosphate Phosphohydrolase 1 (DIPP1/NUDT3) in Complex with 3-Diphosphoinositol 1,2,4,5-Tetrakisphosphate (3-Pp- IP4), Mg and Fluoride Ion
Enzymatic activity of Diphosphoinositol Polyphosphate Phosphohydrolase 1 (DIPP1/NUDT3) in Complex with 3-Diphosphoinositol 1,2,4,5-Tetrakisphosphate (3-Pp- IP4), Mg and Fluoride Ion
All present enzymatic activity of Diphosphoinositol Polyphosphate Phosphohydrolase 1 (DIPP1/NUDT3) in Complex with 3-Diphosphoinositol 1,2,4,5-Tetrakisphosphate (3-Pp- IP4), Mg and Fluoride Ion:
3.6.1.52;
3.6.1.52;
Protein crystallography data
The structure of Diphosphoinositol Polyphosphate Phosphohydrolase 1 (DIPP1/NUDT3) in Complex with 3-Diphosphoinositol 1,2,4,5-Tetrakisphosphate (3-Pp- IP4), Mg and Fluoride Ion, PDB code: 7tn4
was solved by
G.Zong,
H.Wang,
S.B.Shears,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 31.40 / 1.85 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 45.149, 59.743, 62.769, 90, 90, 90 |
| R / Rfree (%) | 18.3 / 21.7 |
Other elements in 7tn4:
The structure of Diphosphoinositol Polyphosphate Phosphohydrolase 1 (DIPP1/NUDT3) in Complex with 3-Diphosphoinositol 1,2,4,5-Tetrakisphosphate (3-Pp- IP4), Mg and Fluoride Ion also contains other interesting chemical elements:
| Fluorine | (F) | 1 atom |
| Magnesium | (Mg) | 3 atoms |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Diphosphoinositol Polyphosphate Phosphohydrolase 1 (DIPP1/NUDT3) in Complex with 3-Diphosphoinositol 1,2,4,5-Tetrakisphosphate (3-Pp- IP4), Mg and Fluoride Ion
(pdb code 7tn4). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 2 binding sites of Chlorine where determined in the Diphosphoinositol Polyphosphate Phosphohydrolase 1 (DIPP1/NUDT3) in Complex with 3-Diphosphoinositol 1,2,4,5-Tetrakisphosphate (3-Pp- IP4), Mg and Fluoride Ion, PDB code: 7tn4:
Jump to Chlorine binding site number: 1; 2;
In total 2 binding sites of Chlorine where determined in the Diphosphoinositol Polyphosphate Phosphohydrolase 1 (DIPP1/NUDT3) in Complex with 3-Diphosphoinositol 1,2,4,5-Tetrakisphosphate (3-Pp- IP4), Mg and Fluoride Ion, PDB code: 7tn4:
Jump to Chlorine binding site number: 1; 2;
Chlorine binding site 1 out of 2 in 7tn4
Go back to
Chlorine binding site 1 out
of 2 in the Diphosphoinositol Polyphosphate Phosphohydrolase 1 (DIPP1/NUDT3) in Complex with 3-Diphosphoinositol 1,2,4,5-Tetrakisphosphate (3-Pp- IP4), Mg and Fluoride Ion
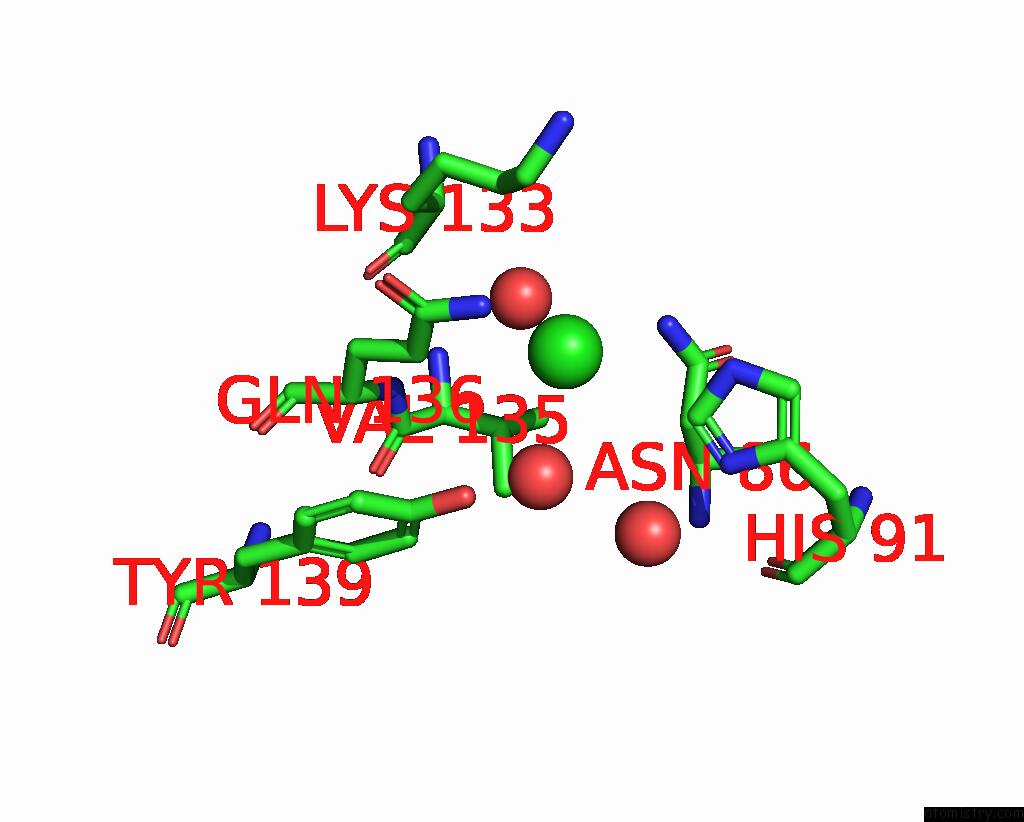
Mono view
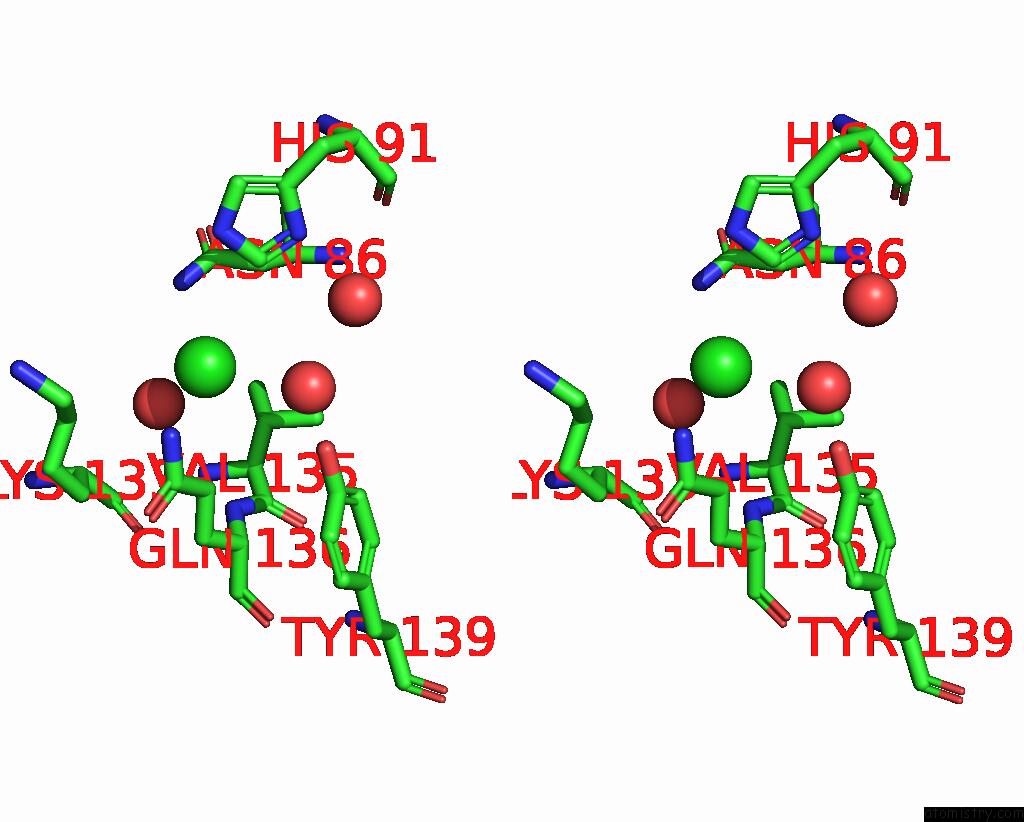
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Diphosphoinositol Polyphosphate Phosphohydrolase 1 (DIPP1/NUDT3) in Complex with 3-Diphosphoinositol 1,2,4,5-Tetrakisphosphate (3-Pp- IP4), Mg and Fluoride Ion within 5.0Å range:
|
Chlorine binding site 2 out of 2 in 7tn4
Go back to
Chlorine binding site 2 out
of 2 in the Diphosphoinositol Polyphosphate Phosphohydrolase 1 (DIPP1/NUDT3) in Complex with 3-Diphosphoinositol 1,2,4,5-Tetrakisphosphate (3-Pp- IP4), Mg and Fluoride Ion

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Diphosphoinositol Polyphosphate Phosphohydrolase 1 (DIPP1/NUDT3) in Complex with 3-Diphosphoinositol 1,2,4,5-Tetrakisphosphate (3-Pp- IP4), Mg and Fluoride Ion within 5.0Å range:
|
Reference:
G.Zong,
S.B.Shears,
H.Wang.
Structural and Catalytic Analyses of the Insp 6 Kinase Activities of Higher Plant Itpks. Faseb J. V. 36 22380 2022.
ISSN: ESSN 1530-6860
PubMed: 35635723
DOI: 10.1096/FJ.202200393R
Page generated: Sun Jul 13 07:31:20 2025
ISSN: ESSN 1530-6860
PubMed: 35635723
DOI: 10.1096/FJ.202200393R
Last articles
Mg in 6WOXMg in 6WOC
Mg in 6WOB
Mg in 6WOE
Mg in 6WOD
Mg in 6WOA
Mg in 6WO9
Mg in 6WO8
Mg in 6WNR
Mg in 6WNQ