Chlorine »
PDB 7tpm-7u1r »
7tz6 »
Chlorine in PDB 7tz6: Structure of Mitochondrial BC1 in Complex with Ck-2-68
Enzymatic activity of Structure of Mitochondrial BC1 in Complex with Ck-2-68
All present enzymatic activity of Structure of Mitochondrial BC1 in Complex with Ck-2-68:
1.10.2.2;
1.10.2.2;
Other elements in 7tz6:
The structure of Structure of Mitochondrial BC1 in Complex with Ck-2-68 also contains other interesting chemical elements:
| Iron | (Fe) | 10 atoms |
| Fluorine | (F) | 6 atoms |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Structure of Mitochondrial BC1 in Complex with Ck-2-68
(pdb code 7tz6). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 2 binding sites of Chlorine where determined in the Structure of Mitochondrial BC1 in Complex with Ck-2-68, PDB code: 7tz6:
Jump to Chlorine binding site number: 1; 2;
In total 2 binding sites of Chlorine where determined in the Structure of Mitochondrial BC1 in Complex with Ck-2-68, PDB code: 7tz6:
Jump to Chlorine binding site number: 1; 2;
Chlorine binding site 1 out of 2 in 7tz6
Go back to
Chlorine binding site 1 out
of 2 in the Structure of Mitochondrial BC1 in Complex with Ck-2-68
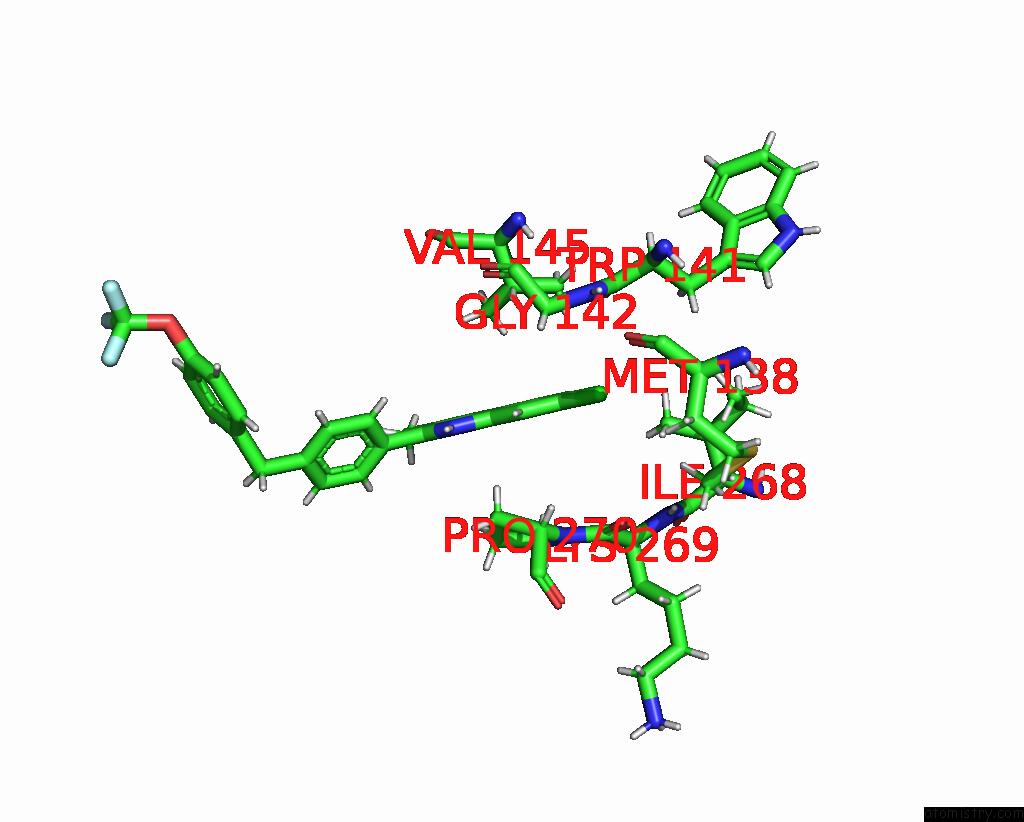
Mono view
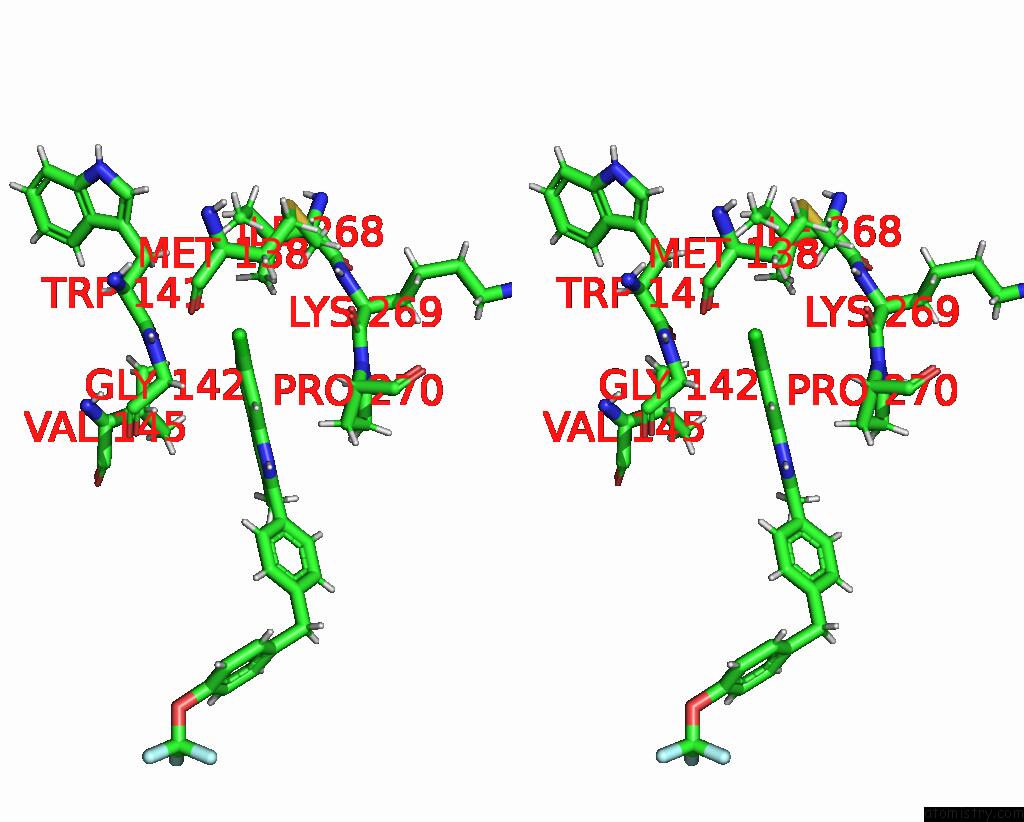
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Structure of Mitochondrial BC1 in Complex with Ck-2-68 within 5.0Å range:
|
Chlorine binding site 2 out of 2 in 7tz6
Go back to
Chlorine binding site 2 out
of 2 in the Structure of Mitochondrial BC1 in Complex with Ck-2-68

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Structure of Mitochondrial BC1 in Complex with Ck-2-68 within 5.0Å range:
|
Reference:
D.Xia,
L.Esser,
F.Zhou.
Conformation Switch of Rieske Isp Subunit Is Revealed By the Crystal Structure of Bacterial Cytochrome BC1 in Complex with Azoxystrobin To Be Published.
Page generated: Sun Jul 13 07:40:35 2025
Last articles
Sr in 3BNQSr in 2QJY
Sr in 2X53
Sr in 2SPT
Sr in 2XRM
Sr in 2WOH
Sr in 2RIO
Sr in 2PN4
Sr in 2QJK
Sr in 2QJP